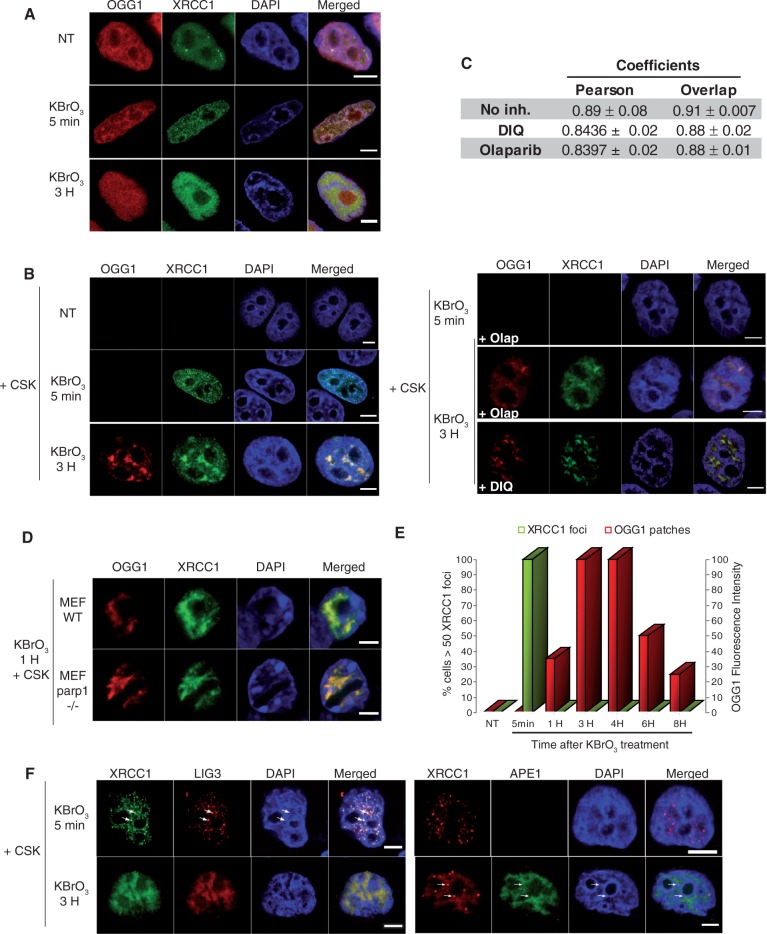
Figure 4.
Distinct subcellular re-localizations of SSBR and BER proteins after global oxidative DNA damage. (A) HeLa cells expressing XRCC1-GFP and OGG1-DsRED were treated with KBrO3 for 30 min, washed and allowed to recover in DMEM for 5 min or 3 h. (B) 5 min or 3H after KBrO3 treatment, in the presence or absence of PARP inhibitors Olaparib or DIQ, cells were washed with CSK buffer and fixed. (C) As indicators of co-localization, Pearson and Overlap correlation coefficients between XRCC1 (green) and OGG1 (red) signals were determined. Values represent the average of coefficients calculated for 10 cells ± SEM. The Kruskal and Wallis statistical test indicates that there is no significant difference in the absence or presence of PARP inhibitors (DIQ or Olaparib). (D) Co-localization of OGG1 and XRCC1 in the CSK resistant fraction in parp1−/− MEF cells after KBrO3 treatment. (E) Kinetics of XRCC1 foci and OGG1 patches formation after KBrO3 treatment. The percentage of cells presenting >50 foci detected without CSK pre-extraction is represented after substracting those present in non-treated cells (green). Fluorescence intensity was used to quantify OGG1-GFP patches after CSK treatment. (F) HeLa cells expressing XRCC1-GFP and LIG3-RFP or APE1-GFP and XRCC1-DsRED monomer were treated with KBrO3 as above and allowed to recover in DMEM for 5 min or 3 h and washed with CSK before fixation. Arrows indicate XRCC1 foci in which LIG3 but not APE1 proteins could be detected. A good co-localization of both proteins with XRCC1 could be observed 3 h after KBrO3 treatment. Scale bar, 5 µm.