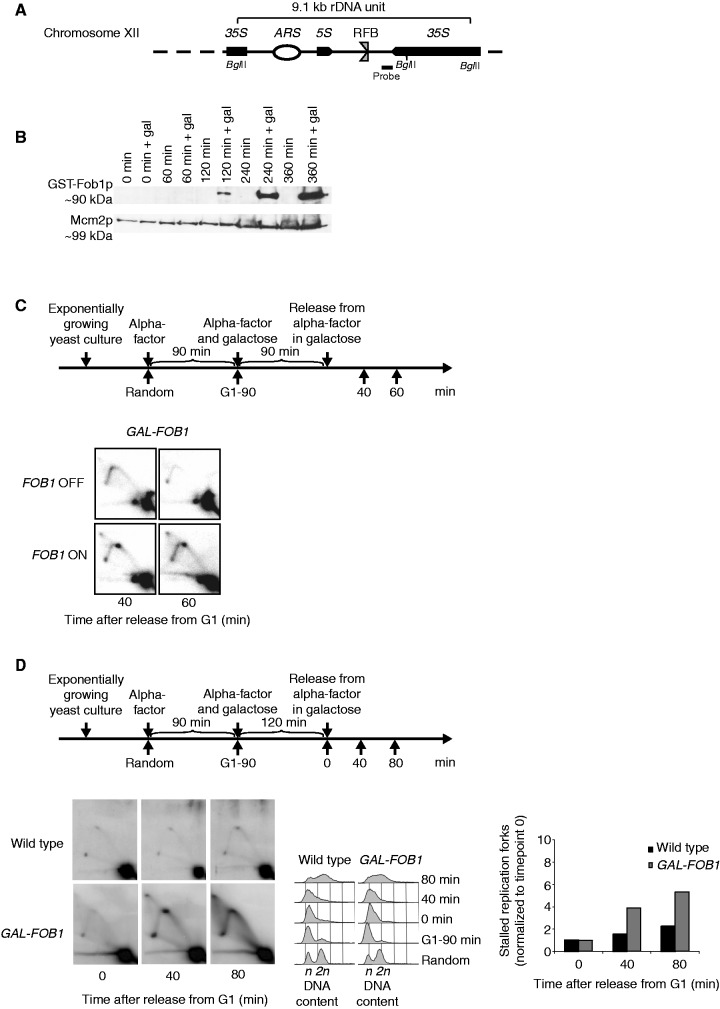
Figure 1.
Verification of the Fob-block system. (A) Schematic illustration of a single 9.1 kb repeat of the rDNA in S. cerevisiae found on Chr. XII. Position of the RFB, origin of DNA replication (ARS), 35S rRNA (35S) and 5S rRNA (5S) genes are indicated. Position of probe used for Southern blotting and BglII cleavage sites used for the 2D DNA gel are also shown. The RFB allows progression of the replication fork in the same direction as 35S rRNA transcription, but not in the opposite direction when Fob1p is bound to the sequence (B) Fob1p expression after different times of galactose induction (0, 60, 120, 240 and 360 min) together with negative controls (non-inducing conditions) on strain LBy-365. (C, upper panel) Outline of yeast culture treatment preceding isolation of DNA for 2D DNA gel analyses. (C, lower panel) 2D DNA gel analysis (strain LBy-413) reveals replication fork stalling in the rDNA (Chr. XII) during inducing conditions (FOB1 ON), but not during non-inducing conditions (FOB1 OFF); see text for details. (D) 2D DNA gel analyses conducted to compare replication fork stalling efficiency at the RFB between a wild-type (LBy-1) strain with endogenous levels of Fob1 and a strain (LBy-413) with overexpression of Fob1. During the experiment, the wild-type strain was kept in glucose medium to give optimal conditions for this strain, whereas LBy-413 was treated as indicated above the 2D gel. Both strains were synchronized. On the right is shown a quantification of replication fork stalling (see ‘Material and Methods’ section for further explanation), and in the middel is shown FACS profiles for the strains. The n and 2 n is DNA content in G1 and G2, respectively.