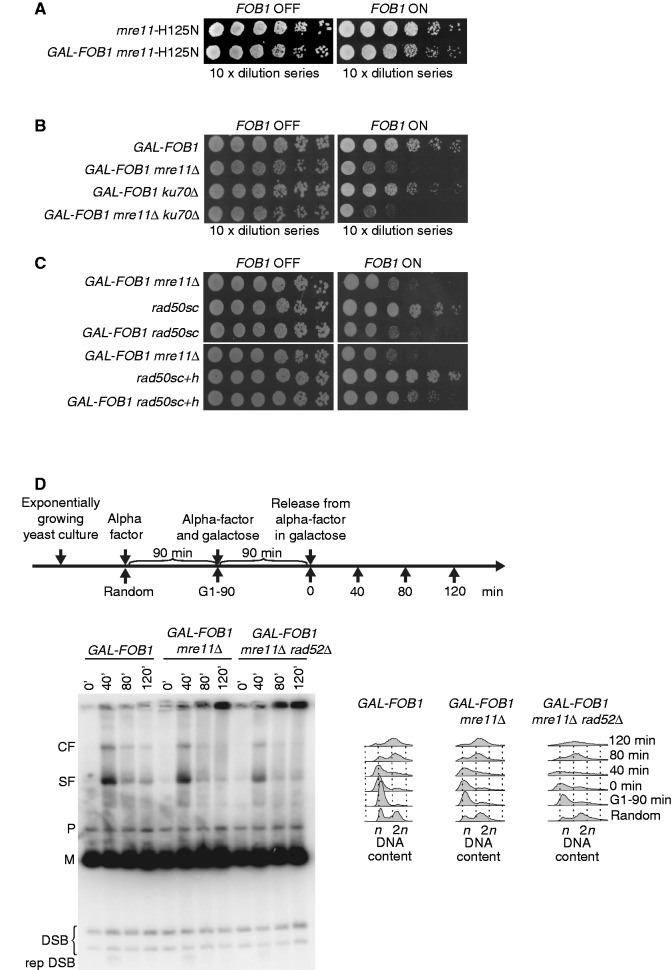
Figure 3.
The MRX complex plays a DSB independent role at RFBs (A) Shown are isogenic strains of mre11-H125N (LBy-623), GAL-FOB1 mre11-H125N (LBy-631). (B) Deletion of KU70 does not suppress the phenotype of GAL-FOB1 mre11 mutant. Shown are isogenic strains of GAL-FOB1 (LBy-413), GAL-FOB1 mre11Δ (LBy-756), GAL-FOB1 ku70Δ (LBy-905) and GAL-FOB1 mre11Δ ku70Δ (LBy-903). (C) Molecular bridging is required by MRX complexes is required at protein–DNA barriers. Shown are isogenic strains of GAL-FOB1 mre11Δ (LBy-756), rad50sc (LBy-1061), GAL-FOB1 rad50sc (LBy-1070), rad50sc+h (LBy-1062) and GAL-FOB1 rad50sc+h (LBy-1071) (D) Absence of Mre11 does not generate more DSBs in the rDNA, but abnormal structures are detected. Shown is a DSB assay for GAL-FOB1 (LBy-413), GAL-FOB1 mre11Δ (LBy-756) and GAL-FOB1 mre11Δ rad52Δ (LBy-680). Digested rDNA units (M), partial digested rDNA units (P), converging forks (CF), stalled forks at RFB (SF), replication independent DSBs (DSB), replication dependent DSB (rep DSB).