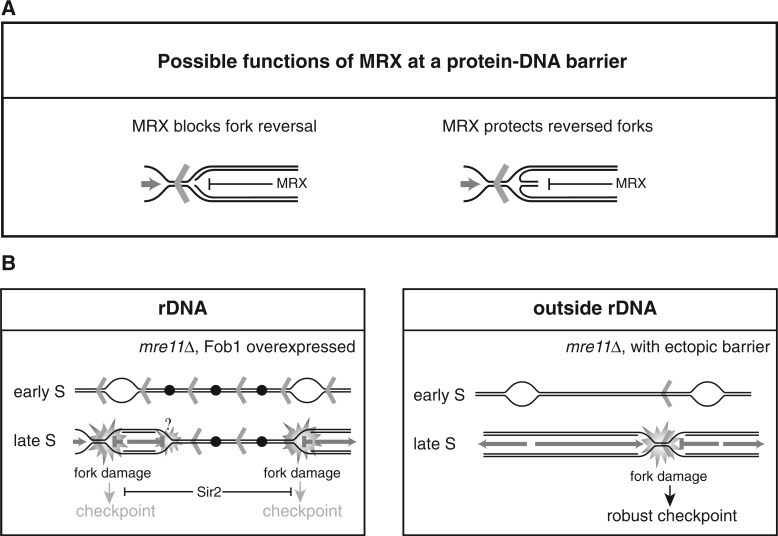
Figure 9.
(A) Suggested functions of the MRX complex on replication fork stalling at a protein–DNA barrier. The MRX complex may hinder fork reversal (left) or protect a reversed fork from further processing (right). (B) Model for the cellular consequences to elevated levels of replication fork stalling in the rDNA (left panel), and for ectopically placed RFBs (right panel) in mre11Δ cells. Fob1 induction generates a higher level of active RFBs in the rDNA, which have detrimental effect on growth when the MRX complex is absent. The growth defect may stem from problems arising directly at the RFBs; however, as replication in the rDNA is challenged by its repetitive nature where unusual structures have the potential to form, fork stalling at other places than the RFB may be a frequent event. Together, this creates a strong need for the MRX complex either to suppress fork reversal or protect reversed forks in the rDNA. When the RFB sequence is placed ectopically, the MRX complex is required at the protein–DNA barrier as the rightward-moving fork is replication competent throughout the ARS606–ARS607 region. Aberrant structures generated in the absence of the MRX complex are checkpoint blind in the rDNA owing to the presence of Sir2, whereas ectopically placed RFBs provoke a checkpoint signal in the absence of the MRX complex. Open bubbles represent active origins, whereas black dots represent inactive origins. Red arrowheads represent Fob1-bound RFB sequences.