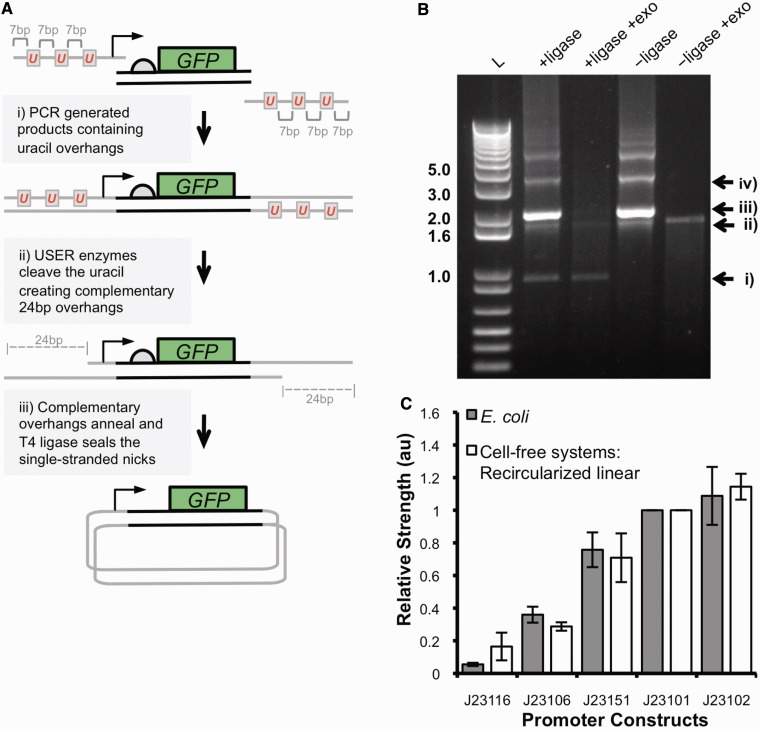
Figure 5.
(A) A schematic of the USER–ligase assembly strategy. (i) PCR is performed with PfuTurbo Cx DNA polymerase to amplify DNA regulatory elements upstream of a GFP coding region with overhangs containing three uracil nucleotides 7-bp apart. The PCR product is purified, and a USER–ligase reaction is performed. (ii) The USER enzymes excise uracil nucleotides creating 24-bp complementary single-stranded overhangs that anneal. (iii) T4 DNA ligase forms a phosphodiester at the single-stranded nicks of the annealed overhangs, forming closed-circular DNA. (B) Products and digestion of a USER–ligase and USER-only reaction analysed on GelRed strained agarose gel. L: 1 kb plus DNA ladder; + ligase and − ligase indicate the presence or absence of T4 DNA ligase in the USER reaction; + exo indicates where digestion by exonuclease III has been performed. Enzymatic digestions identify the products of the USER–ligase reaction as follows: (i) closed-circular DNA, (ii) linear DNA products, (iii) annealed DNA containing single-stranded nicks and (iv) concatemers from ligation of multiple PCR products. + ligase and − ligase lanes contain 300 ng of DNA products. Enzymatic digestion with exonuclease III contained 300 ng of DNA products. (C) The relative promoter strength of five minimal σ70 constitutive promoters was calculated from cell-free systems using 1 µg of USER–ligase products for each reaction and fluorescence measured. The relative promoter strength was calculated from the rate of fluorescence increase in the maximal phase of GFP expression, 30–45 min after initiation, and normalized to the strength of the J23101 promoter. The data represent the mean +/− the SD of three independent reactions. The relative promoter strength was compared with the relative strengths calculated from experiments in E. coli.