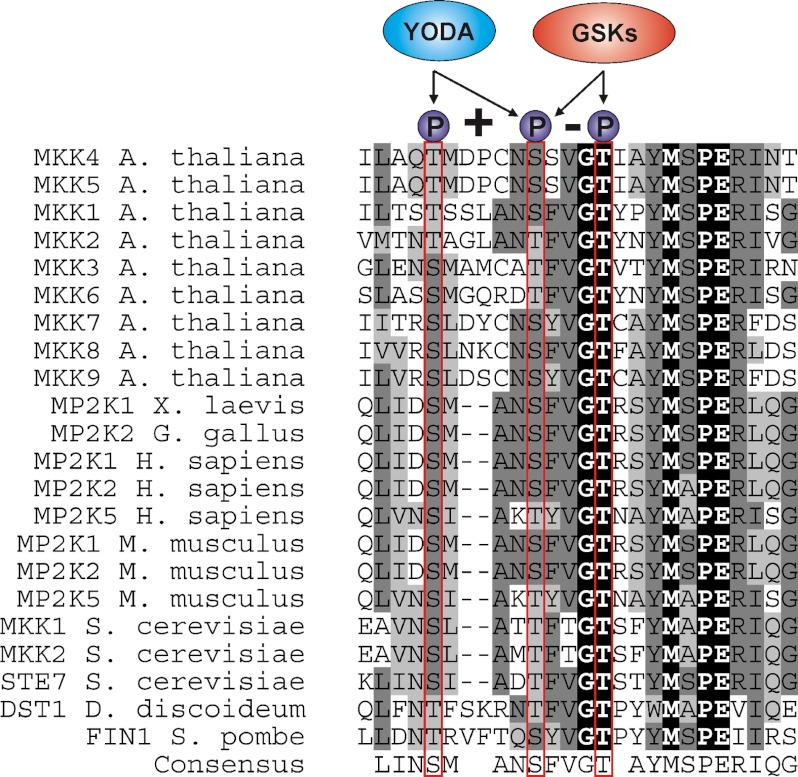
FIGURE 7.
Amino acid alignment of MAPKKs from different eukaryotes. Dashes indicate gaps introduced to maximize alignment of conserved residues. Residues corresponding to Thr-224, Ser-230, and Thr-234 are marked in red. X. laevis, Xenopus laevis; G. gallus, Gallus gallus; H. sapiens, Homo sapiens; M. musculus, Mus musculus; S. cerevisiae, Saccharomyces cerevisiae; D. discoideum, Dictyostelium discoideum; S. pombe, Schizosaccharomyces pombe.