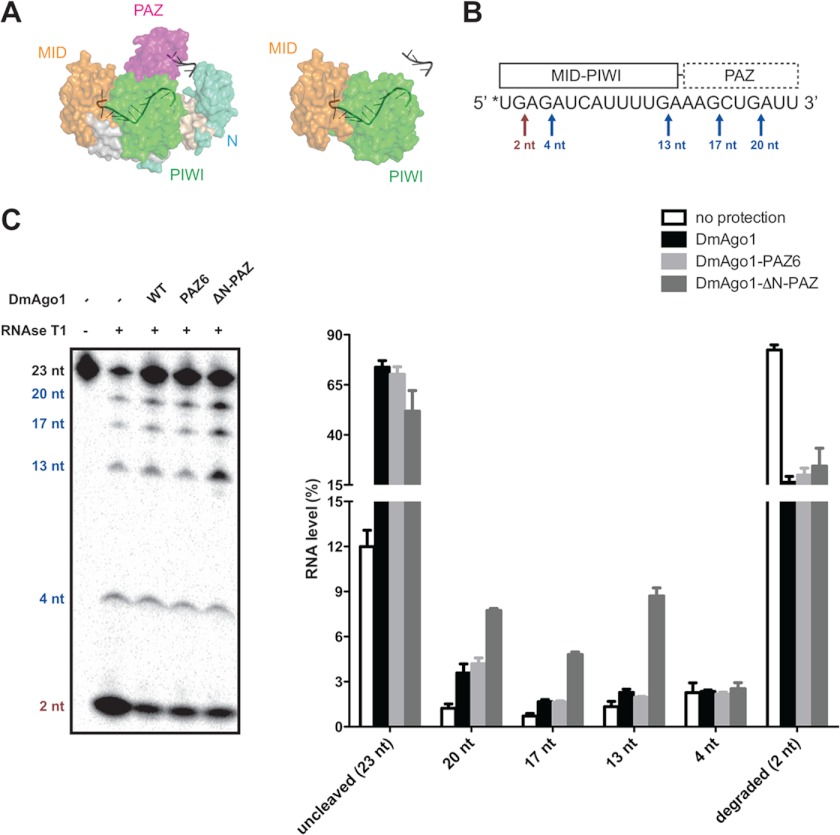
FIGURE 2.
Argonaute N-PAZ lobe protects the 3′ end of guide RNA. A, surface representation of T. thermophilus Argonaute structure in complex with 21-nt DNA (Protein Data Bank code 3DLH). Light blue, N; magenta, PAZ; yellow, MID; green, PIWI; gray, bound DNA (only nucleotides 1–11 and 18–21 are traced in structure). Wild type is shown on the left with theoretical deletion of N-PAZ-lobe on the right. B, schematic of RNase T1 protection by DmAgo1 variants: 5′ radioabeled (*) bantam miRNA (23 nt) is depicted with known interactions to the PAZ, MID, and PIWI domains. RNase T1 cleavage sites, which occur 3′ of guanosine residues, are marked with arrows. The red and blue arrows indicate cleavage sites protected by the MID-PIWI lobe and N-PAZ lobe, respectively. C, DmAgo1-miRNA complex subject to RNase T1 protection assay. 5′-Labeled bantam miRNA was incubated with excess amount of purified DmAgo1 variants and was subjected to partial digestion by RNase T1. The amount of each RNA species was normalized with respect to total RNA.