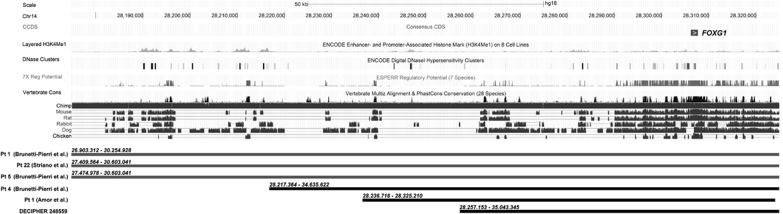
Figure 1.
UCSC genome browser (NCBI36/hg18) schematic view of reported 14q12 duplications encompassing FOXG1 gene. Histone modifications from the ENCODE project7 in eight cells lines (GM12878, H1-hESC, HMEC, HSMM, HUVEC, K562, NHEK and NHLF) indicate the presence of regulatory elements upstream FOXG1 gene. Monomethylation of histone 3 at lysine1 (H3K4me1) is often found near regulatory elements, whereas the presence of DNaseI hypersensitive clusters reflect genomic regions containing regulatory informations. Potential regulatory elements identified with the ESPERR (Evolutionary and Sequence Pattern Extraction through Reduced Representations) computational method and evolutionary conservation are also shown. Grey bars represent duplications associated with an epileptic phenotype; black bars represent duplication identified in non-epileptic patients.