Abstract
Homogeneous Proto-Slavic genetic substrate and/or extensive mixing after World War II were suggested to explain homogeneity of contemporary Polish paternal lineages. Alternatively, Polish local populations might have displayed pre-war genetic heterogeneity owing to genetic drift and/or gene flow with neighbouring populations. Although sharp genetic discontinuity along the political border between Poland and Germany indisputably results from war-mediated resettlements and homogenisation, it remained unknown whether Y-chromosomal diversity in ethnically/linguistically defined populations was clinal or discontinuous before the war. In order to answer these questions and elucidate early Slavic migrations, 1156 individuals from several Slavic and German populations were analysed, including Polish pre-war regional populations and an autochthonous Slavic population from Germany. Y chromosomes were assigned to 39 haplogroups and genotyped for 19 STRs. Genetic distances revealed similar degree of differentiation of Slavic-speaking pre-war populations from German populations irrespective of duration and intensity of contacts with German speakers. Admixture estimates showed minor Slavic paternal ancestry (∼20%) in modern eastern Germans and hardly detectable German paternal ancestry in Slavs neighbouring German populations for centuries. BATWING analysis of isolated Slavic populations revealed that their divergence was preceded by rapid demographic growth, undermining theory that Slavic expansion was primarily linguistic rather than population spread. Polish pre-war regional populations showed within-group heterogeneity and lower STR variation within R-M17 subclades compared with modern populations, which might have been homogenised by war resettlements. Our results suggest that genetic studies on early human history in the Vistula and Oder basins should rely on reconstructed pre-war rather than modern populations.
Keywords: Y chromosome, demography, admixture, Slavs, Poland, Germany
Introduction
The male genetic landscape of the European continent has been shown to be clinal and influenced primarily by geography rather than by language.1 One of the most outstanding phenomena in the Y-chromosomal diversity in Europe concerns the population of Poland, which reveals geographic homogeneity of Y-chromosomal lineages in spite of a relatively large geographic area seized by the Polish state.2 Moreover, a sharp genetic border has been identified between paternal lineages of neighbouring Poland and Germany, which strictly follows a political border between the two countries.3 Massive human resettlements during and shortly after the World War II (WWII), involving millions of Poles and Germans, have been proposed as an explanation for the observed phenomena.2, 3 Thus, it was possible that the local Polish populations formed after the early Slavic migrations displayed genetic heterogeneity before the war owing to genetic drift and/or gene flow with neighbouring populations. It has been also suggested that the revealed homogeneity of Polish paternal lineages existed already before the war owing to a common genetic substrate inherited from the ancestral Slavic population after the Slavs' early medieval expansion in Europe.2
From the linguistic point of view, western Slavic dialects are classified as Czech/Slovak, Lusatian and Lekhitic; the Lekhitic branch is further divided into Polish, Pomeranian and Polabian.4 Nowadays, among the western Slavs, only Polish and Czech/Slovak dialects have evolved into fully viable languages with millions of speakers. Lusatian is spoken by 66 000 Sorbs inhabiting southeastern Germany, down from 166 000 speakers in the late 19th century.5 Present-day Pomeranian comprises 53 000 speakers of Kashubian in northern Poland,6 although roughly half a million people in Poland claim Kashubian and half Kashubian ancestry.7 While Slavists classify Kashubian as a separate Slavic language,4 the vast majority of Kashubes declare Polish ethnicity.6 Polabian was spoken until the 18th century in what is now northeastern Germany.8 The Polish linguistic area is further subdivided into four dialectal groups, roughly corresponding to early Slavic tribal division: Greater Polish, Lesser Polish, Silesian and the most linguistically divergent Masovian.9
There exists an opinion among academics that ‘the Slavic ethnogenesis remains a major, if not the most important, topic in the historiography of Eastern Europe'.10 Most of the current knowledge on this subject results from indirect evidence based on linguistics, archaeology and anthropology, including, since recently, molecular genetics.11 The changes seen in the 5th–6th centuries in eastern Europe are explained either in terms of a demographic expansion of the Slavic people, carrying with them their genes, customs and language, or as a primarily linguistic spread with only minor contribution of migration.12
We used high-resolution typing of Y-chromosomal binary and microsatellite markers first to test for male genetic structure in the Polish population before massive human resettlements in the mid-20th century, and second to verify if the observed present-day genetic differentiation between the Polish and German paternal lineages is a direct consequence of the WWII or it has rather resulted from a genetic barrier between peoples with distinct linguistic backgrounds. The study further focuses on providing an answer to the origin of the expansion of the Slavic language in early medieval Europe. For the purpose of our investigation, we have sampled three pre-WWII Polish regional populations, three modern German populations (including the Slavic-speaking Sorbs) and a modern population of Slovakia.
Materials and methods
A total of 1156 individuals were analysed in the present study, including 520 unrelated males descending directly from pre-WWII native inhabitants of three distinct ethnolinguistic regions of Poland: Kaszuby (Kashubian-speaking region, n=204), Kociewie (Greater Polish-speaking region, n=158) and Kurpie (Masovian-speaking region, n=158). Inhabitants of the Kurpie region trace their origin to Masovian peasants who since the 16th century colonised forests between Masovia and Prussia, and were subjected to some degree of geographic and cultural isolation.9 The Kashubian samples were additionally assigned to three different dialects:9 northern (n=70), central (n=93) and southern (n=41). As genetic distances revealed the three Kashubian subpopulations to be genetically undistinguishable (data not shown), they were treated in many subsequent analyses as one population. Only individuals whose ancestors were born in villages and inhabiting the studied areas for at least three generations in paternal lineages were selected for the study. In addition, a sample set from Germany comprised Sorbs from Lusatia (Upper Sorbian speakers, n=123) and Germans from Mecklenburg (northeastern Germany, n=131) and western Bavaria (southwestern Germany, n=218). Finally, DNA samples from western Slovakia (n=164), used previously in a comprehensive analysis of Y-STR variation in the Slavic populations,11 were also included in the study. The studied populations and their linguistic background are summarised in Table 1, while their geographic locations on an ethnolinguistic map of central Europe in the early 20th century are shown in Supplementary Figure S1.
Table 1. Linguistic affiliations, Y-STR MPD and WIMP values (±SD), and surname distributions for the analysed populations.
| Population | Linguistic affiliation | MPD | WIMP | Slavic vs German surnames |
|---|---|---|---|---|
| Kaszuby (n=204) | W Slavic, Pomeranian, Kashubian | 9.26±4.27 | 5.07±1.29 | 0.681: 0.250 |
| Kociewie (n=158) | W Slavic, Polish, Greater Polish | 9.30±4.30 | 5.23±1.15 | 0.791: 0.177 |
| Kurpie (n=158) | W Slavic, Polish, Masovian | 9.32±4.30 | 4.70±1.15 | 0.873: 0.089 |
| Lusatia (n=123) | W Slavic, Lusatian, Upper Sorbian | 8.24±3.85 | 4.23±1.31 | — |
| Slovakia (n=164) | W Slavic, Czech/Slovak, W Slovak | 9.83±4.52 | 4.92±1.02 | — |
| Mecklenburg (n=131) | German | 10.04±4.62 | 5.19±0.82 | — |
| Bavaria (n=218) | German | 10.43±4.77 | 5.50±0.75 | — |
Abbreviations: MPD, mean pairwise difference; WIMP, weighted mean intralineage mean pairwise difference.
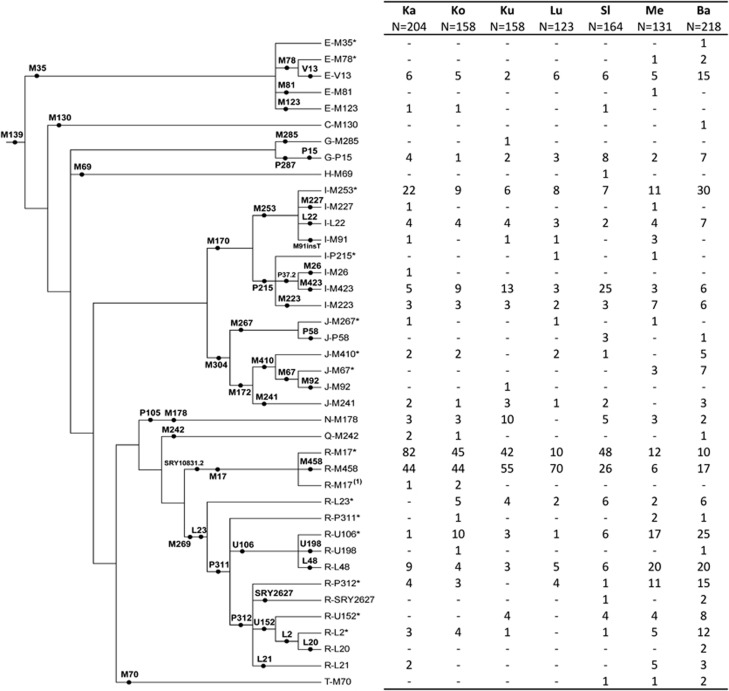
Two multiplex PCRs were utilised to genotype a total of 19 Y-STRs, including 17 STRs present in the commercially available AmpFlSTR Yfiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA). The second multiplex comprised two additional Y-STRs: DYS388 and DYS426, as well as six biallelic markers, displaying amplified fragment length polymorphism: A-M91, BT-M139, B-M60, M-M186, O-M175 and R-M17.13 As the Yfiler kit amplifies two DYS385 loci simultaneously avoiding their discrimination, DYS385 was excluded from all the analyses performed, providing a total of 17 Y-STRs (including DYS388 and DYS426) for inferences. Other Y-SNPs were genotyped individually with the use of pre-designed TaqMan assays with previously published primer sequences.14 Their phylogenetic relationship is shown in Figure 1.
Figure 1.
Phylogenetic relationship and frequencies of Y-chromosomal haplogroups in the studied populations. Ka Kaszuby; Ko Kociewie; Ku Kurpie; Lu Lusatia; Sl Slovakia; Me Mecklenburg; Ba Bavaria. (1) R-M17-derived samples with unknown M458 status owing to permanent lack of PCR product, which the most likely resulted from deletion of the M458 locus, located in very close proximity to the DYS448 marker (independent deletions of DYS448 have been described within different haplogroups45 and two out of the three samples with unknown R-M458 genotypes possess DYS448 null alleles).
Observed haplogroup frequencies were employed to calculate a matrix of pairwise FST values. Y-STR haplotypes were used to obtain ΦST and RST molecular distances. Calculations of genetic distances, estimations of corresponding P-values based on 10 000 permutations and analysis of molecular variance (AMOVA) were performed with the use of Arlequin 3.1 software.15 In order to thoroughly explore the Y chromosome distribution in the Polish population before and after the WWII, our data were compared with 7-STR haplotypes published for a pre-WWII southern Polish population from the Lesser Polish-speaking regions of Podhale and Sądecczyzna (n=140)16 and for a number of modern Polish populations,16, 17, 18 including Kaszuby (n=142) and Podhale and Sądecczyzna (n=226). Multidimensional scaling (MDS) based on linearised distances19 was carried out with the use of STATISTICA 9.1 software (StatSoft, Tulsa, OK, USA). Network 4.6 software (Fluxus Technology, Clare, UK) was applied to build a median-joining network20 of Y-STR haplotypes with a maximum parsimony option.21 Mean pairwise differences (MPDs) within populations based on the 17-STR haplotypes and the weighted mean intralineage MPDs (WIMPs) were calculated as previously described.22 STR variation within chosen haplogroups was assessed by genetic variance (VP)23 and by average squared difference in the number of repeats between all chromosomes and a median haplotype, averaged over microsatellite loci (ASD0).24
The pre-WWII Polish samples were additionally divided into three subgroups, depending on surnames of the tested individuals. The first group comprised individuals carrying surnames with roots revealing Slavic/eastern European etymology or origin. Accordingly, males with surname roots indicating German/western European etymology or origin were included in the second group. The third group contained surnames with unclear or hybrid etymology. For each surname, the assignment was based on linguistic analysis provided in etymological dictionaries.25, 26, 27
BATWING28 was used to assess time of demographic expansion and split of the populations of Kaszuby and Lusatia. Time of start of demographic expansion, growth rate and time of population split were estimated using a model of exponential growth from a constant-size ancestral population. Observed mutation rates for each marker were used in the analysis.29 Y-STR mutation data published in the Y Chromosome Haplotype Reference Database30 and in the literature29, 31 were used to set mutation rate priors as provided in Supplementary Table S1. An initial effective population size and growth rate were given priors of gamma(1.1,0.0001) and gamma(1.01,1), respectively, in order to cover very wide ranges of possible values.32 Maximally uninformative uniform priors were set for dates of the expansion start and population split. SNP information was integrated for the phylogenetic reconstruction, but it was not considered for posterior estimates. A total of 10 million Markov chain Monte Carlo (MCMC) samples were collected: the first 5 million were rejected as burn-in and the remaining 5 million were used for inference. BATWING convergence was assessed from two independent runs with different seeds with the use of Gelman and Rubin's convergence diagnostic available in the CODA package for R.33, 34 In order to put the BATWING results in a historical time scale, a male generation interval of 31 years35 was used.
Populations speaking Sorbian and Kashubian, linguistically the most closely related to extinct Slavic dialects spoken in the past in present-day eastern Germany, were used to assess Slavic ancestry in the eastern German Y-chromosomal pool. In addition, German admixture was assessed in genetic outliers detected in the MDS analysis, that is, the Sorbs and Kashubes, with the Greater Polish-speaking population of Kociewie as the parental population (the Greater Polish dialects directly neighbour the Kaszuby region and share linguistic similarities with the Lusatian dialects9). For haplogroup data, genetic admixture estimators based on allele frequencies were assessed. An mR estimator comparing directly haplogroup frequencies was computed with the use of Admix 2.0.36 A maximum likelihood approach-based mW estimator considering an effect of genetic drift in admixed and parental populations was obtained with the aid of Leadmix software.37 As the overwhelming majority of Y-STR haplotypes were singletons specific to only one population, in case of STR data, an mY estimator taking into account molecular distances between haplotypes rather than haplotype frequencies was computed with the use of Admix 2.0. In order to eliminate likely haplotype homoplasy, SNP phylogeny was integrated into STR information, weighting biallelic mutations 1000-fold higher than STR mutations.38 The molecular relationship between haplotypes was defined as the sum of squared differences in allele sizes.38
Results
A total of 39 different haplogroups have been detected in the studied sample set (Figure 1), including an insertion polymorphism at M91 (M91insT with a stretch of 10 thymidines) previously observed in two individuals from a large worldwide sample set.39 No derived alleles at R-M153 (a subclade of R-P312) and R-M222 (a subclade of R-L21) have been detected. Genotyping results for all 1156 individuals are provided in Supplementary Table S2.
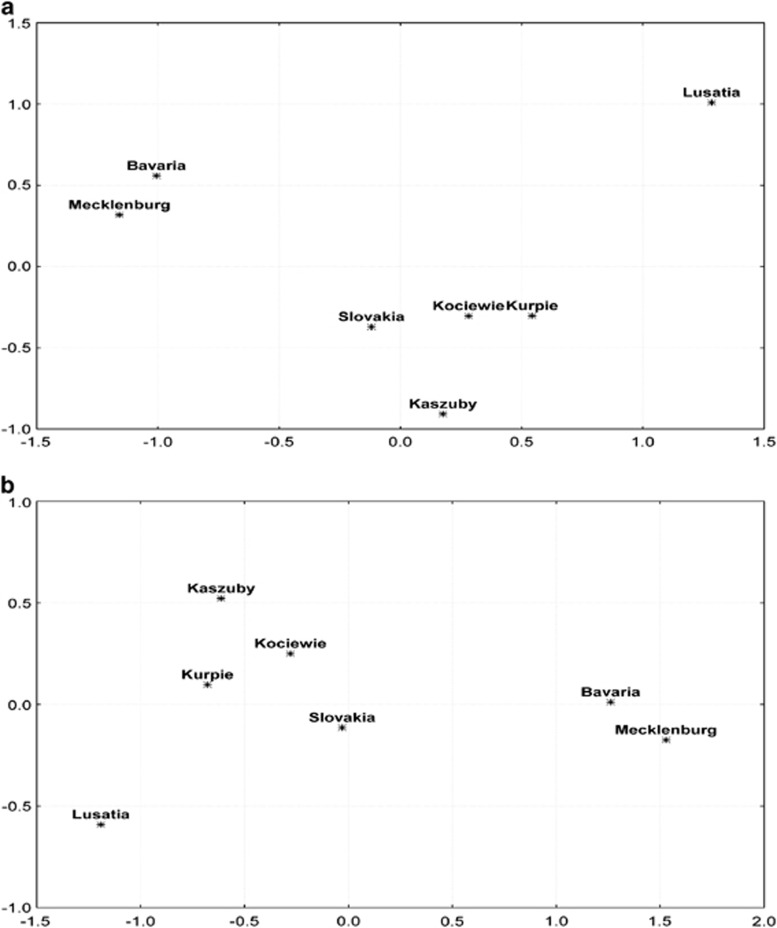
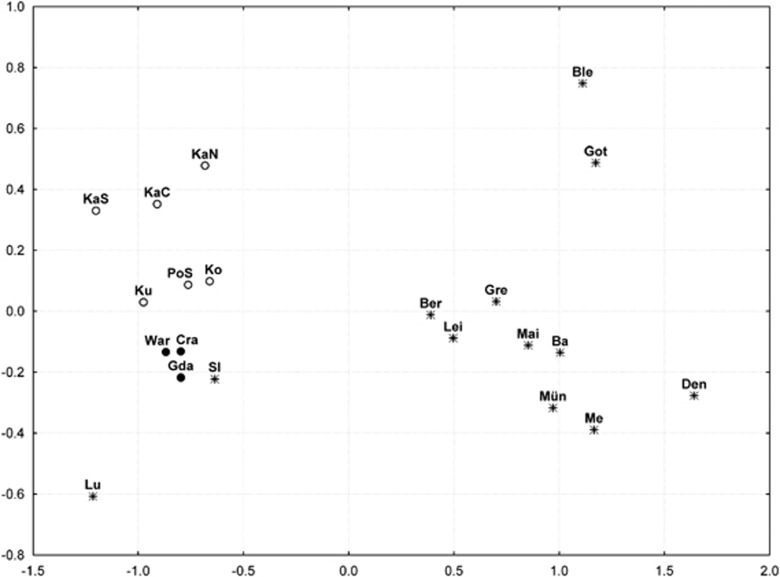
AMOVA in the studied populations revealed statistically significant support for two linguistically defined groups of populations in both haplogroup and haplotype distributions (Table 2). It also detected statistically significant genetic differentiation for both haplogroups and haplotypes in three Polish pre-WWII regional populations (Table 2). The AMOVA revealed small but statistically significant genetic differentiation between the Polish pre-war and modern populations (Table 2). When both groups of populations were tested for genetic structure separately, only the modern Polish regional samples showed genetic homogeneity (Table 2). Regional differentiation of 10-STR haplotypes in the pre-WWII populations was retained even if the most linguistically distinct Kashubian speakers were excluded from the analysis (RST=0.00899, P=0.01505; data not shown). Comparison of Y chromosomes associated with etymologically Slavic and German surnames (with frequencies provided in Table 1) did not reveal genetic differentiation within any of the three Polish regional populations for all three (FST, ΦST and RST) genetic distances. Moreover, the German surname-related Y chromosomes were comparably distant from Bavaria and Mecklenburg as the ones associated with the Slavic surnames (Supplementary Figure S2). MDS of pairwise genetic distances showed a clear-cut differentiation between German and Slavic samples (Figure 2). In addition, the MDS analysis revealed the pre-WWII populations from northern, central and southern Poland to be moderately scattered in the plot, on the contrary to modern Polish regional samples, which formed a very tight, homogeneous cluster (Figure 3).
Table 2. AMOVA results for the studied populations (Hg=39 Y-SNP subclades; Ht17=17 Y-STRs) and for previously published data for Polish pre-war and modern populations (Ht7=7 Y-STRs) (Roewer et al;17 Woźniak et al 16, 18).
| Tested structure | Markers | Statistics | P-value | Percentage of variation |
|---|---|---|---|---|
| 2 groups: 5 Slavic populations vs 2 German populations (this study) | Hg | FCT=0.05715 | 0.04812 | 5.72 |
| FSC=0.03344 | 0.00000 | 3.15 | ||
| FST=0.08868 | 0.00000 | 91.13 | ||
| Ht17, ΦST | ΦCT=0.06669 | 0.05059 | 6.67 | |
| ΦSC=0.00902 | 0.00000 | 0.84 | ||
| ΦST=0.07510 | 0.00000 | 92.49 | ||
| Ht17, RST | RCT=0.10529 | 0.04861 | 10.53 | |
| RSC=0.00940 | 0.00000 | 0.84 | ||
| RST=0.11370 | 0.00000 | 88.63 | ||
| 1 group: 3 Polish pre-war populations (this study) | Hg | FST=0.01356 | 0.00109 | 1.36 |
| Ht17, ΦST | ΦST=0.00246 | 0.06693 | 0.25 | |
| Ht17, RST | RST=0.00749 | 0.01198 | 0.75 | |
| 2 groups: Polish pre-wara vs Polish modernb populations | Ht7, ΦST | ΦCT=0.00157 | 0.01287 | 0.16 |
| ΦSC=0.00158 | 0.03426 | 0.16 | ||
| ΦST=0.00314 | 0.00376 | 99.69 | ||
| Ht7, RST | RCT=0.00201 | 0.03228 | 0.20 | |
| RSC=0.00153 | 0.12337 | 0.15 | ||
| RST=0.00354 | 0.04614 | 99.65 | ||
| 1 group: Polish pre-wara populations | Ht7, ΦST | ΦST=0.00460 | 0.01713 | 0.46 |
| Ht7, RST | RST=0.00688 | 0.03475 | 0.69 | |
| 1 group: Polish modernb populations | Ht7, ΦST | ΦST=0.00047 | 0.26792 | 0.05 |
| Ht7, RST | RST=−0.00042 | 0.56960 | −0.04 |
Abbreviation: AMOVA, analysis of molecular variance.
Polish pre-war populations: Kaszuby (north, centre and south), Kociewie, Kurpie (this study), Podhale and Sądecczyzna (Woźniak et al16).
Figure 2.
MDS analysis of (a) FST values for Y-chromosomal haplogroups and (b) ΦST values for 17-locus Y-STR haplotypes observed in the studied populations.
Figure 3.
MDS analysis based on ΦST distances for 7-locus Y-STR haplotypes observed in the studied populations compared with data published for 12 Slavic and Germanic populations.16, 17 Filled circles indicate modern populations from northern (Gda Gdansk), central (War Warsaw) and southern Poland (Cra Cracow). Empty circles indicate pre-WWII populations from northern (KaN, KaC, KaS northern, central, southern Kaszuby; Ko Kociewie), central (Ku Kurpie) and southern Poland (PoS). Other Slavic populations: Lu Lusatia; Sl western Slovakia. German populations: Me Mecklenburg; Ba western Bavaria; Gre Greifswald; Ber Berlin; Lei Leipzig; Mai Mainz; Mün Münster. Other Germanic populations: Den Denmark; Got Gotland (Sweden); Ble Blekinge (Sweden).
The MPD and WIMP values did not reveal significant reduction in Y-chromosomal diversity in populations with differential degree of cultural and/or geographic isolation, that is, Kaszuby, Lusatia and Kurpie (Table 1). In order to check for the effect of sampling pre-WWII populations on STR variation, genetic variance (VP) and average squared difference (ASD0) were assessed within the most common haplogroups found in the studied Slavic populations: R-M17*(xM458) and R-M458. Both parameters reached lower values in the native pre-WWII populations of the Vistula and Oder basins in comparison with the modern Polish population studied by Underhill et al.40 A value comparable to the modern Poles was obtained only in the case of ASD0 in the R-M17*(xM458) chromosomes from Kaszuby (Table 3). A median-joining network of our R-M17*(xM458) 17-STR haplotypes revealed a clearly separated cluster of Y chromosomes, involving as many as 22 individuals from Kaszuby, as well as several individuals from other Slavic populations (Supplementary Figure S3). The observed cluster is likely to represent an unknown R-M17 subclade and explains the high ASD0 value in haplogroup R-M17*(xM458) among the Kashubes.
Table 3. VP and ASD0 for 17 Y-STRs in haplogroups R-M17*(xM458) and R-M458 in native pre-war regional populations of the Vistula and Oder basins (this study) and in the modern Polish population, studied by Underhill et al40.
| R-M17*(xM458) | R-M458 | |||||
|---|---|---|---|---|---|---|
| Population | n | VP | ASD0 | n | VP | ASD0 |
| Kaszuby | 82 | 0.327 | 0.454 | 44 | 0.160 | 0.170 |
| Kociewie | 45 | 0.334 | 0.413 | 44 | 0.203 | 0.233 |
| Kurpie | 42 | 0.324 | 0.369 | 55 | 0.173 | 0.206 |
| Lusatia | 10 | 0.168 | 0.206 | 70 | 0.176 | 0.209 |
| Poland (modern) | 21 | 0.424 | 0.462 | 29 | 0.223 | 0.262 |
Abbreviations: ASD0, average squared difference; VP, genetic variance.
BATWING of the Slavic populations of Kaszuby and Lusatia provided convergent MCMC chains with unimodal distribution and revealed that their divergence took place 1.7 kya (95% confidence intervals: 1.4–2.1 kya) and was preceded by 0.6 ky of demographic expansion with a 4.2% growth rate (Table 4).
Table 4. Times of demographic expansion and split for Y chromosomes from the populations of Kaszuby and Lusatia.
| Parameter | Modal value with 95% CI |
|---|---|
| Time to the most recent common ancestor | 20.0 kya (16.2–29.3) |
| Expansion start | 2.4 kya (1.8–3.2) |
| Growth rate | 4.2% (3.1–6.4%) |
| Split | 1.7 kya (1.4–2.1) |
| Time between the expansion start and the split | 0.6 ky (0.1–1.4) |
Abbreviation: CI, confidence interval.
As both the Sorbs and Kashubes are historically the most closely related to the extinct Slavic tribes of eastern Germany and none directly contributed to the modern German population of Mecklenburg, it was assumed that the population of Mecklenburg resulted from admixture of western German (Bavarian as a proxy), Sorbian and Kashubian populations. All the ancestry estimates were the highest for the western German population (Supplementary Table S3). On the other hand, admixture analysis failed to detect considerable German ancestry in paternal lineages of genetic outliers detected in the MDS analysis, that is, the Sorbs and Kashubes (Supplementary Table S4). After inclusion of data from German regional populations studied by Kayser et al,3 the Slavic (Sorbian or Kashubian) ancestry estimates mR, mW and mY for the pooled eastern German populations (n=678) in comparison with the pooled western German populations (n=886) ranged from 0.182 to 0.261.
Discussion
Most molecular anthropological studies concerning early human history in Central Europe29, 40, 41 exploit previously observed geographic homogeneity of Polish paternal lineages.2 Although it was suggested that the homogeneous Polish Y-chromosomal gene pool was formed very recently after the massive human resettlements linked to the WWII,2 a previous study on a southern Polish population failed to detect genetic differences between pre-WWII and post-WWII Y chromosomes in the region.16 However, it should be noted that the studied region did not experience massive population exchange and its post-WWII settlers originated mainly in the neighbouring areas.16 The same authors studied a modern population of Kaszuby, the most linguistically distinct ethnic group among modern Poles, and no genetic differentiation within the Polish population was found.18 Our results are based on pre-WWII regional populations from four out of five main Polish linguistic/dialectal groups (Kashubian, Masovian, Greater Polish and Lesser Polish), and demonstrate for the first time that the Polish paternal lineages were unevenly distributed within the country before the forced resettlements of millions of people during and shortly after the WWII. Small but statistically significant differentiation between the pre-WWII and modern populations is particularly remarkable taking into account the fact that modern Polish regional samples comprise varying ratios of pre-WWII inhabitants and post-WWII settlers. The observed heterogeneity suggests that precautions should be taken in order to collect representative population samples from Poland for evolutionary studies, as well as for forensic purposes in case of statistical evaluation of genetic evidence concerning regions densely populated by native pre-WWII inhabitants.
Alternatively, the observed substructure could result from the fact that our pre-WWII samples originated in rural areas that were less likely to be influenced by migrations than large cities,32 whereas Ploski et al2 revealed geographic homogeneity of Y-chromosomal lineages in general populations of several Polish regions. However, it should be noted that WWII-mediated resettlements involved both urban and rural populations. The study by Woźniak et al18 on the modern population of Kaszuby from villages and small towns did not detect its distinctiveness from other modern Polish regional samples, which may be owing to the fact that in 1950, the post-WWII settlers constituted as many as 36.7% of inhabitants of an area roughly corresponding to the regions of Kaszuby and Kociewie42 (in case of populations studied by Ploski et al,2 in 1950, the share of post-WWII settlers ranged from 6.8% in the Cracow region up to 93.8% in the Wroclaw region42) and discards rural origin of our pre-WWII Polish regional populations as the main reason for the detected substructure.
Parameters measuring STR variation within Y-chromosomal haplogroups are commonly used for dating of SNP mutations in order to draw conclusions about origins and history of human populations.23, 24 Underhill et al40 observed the highest genetic diversity in Europe for R-M17*(xM458) and R-M458 subclades in the Vistula and Oder basins, which correspond roughly to the present-day territory of Poland. We examined Y-STR variation within the two subclades in pre-WWII Polish regional populations of the Vistula basin (Kurpie, Kociewie and Kaszuby) and in a native population of the Oder–Elbe basin borderland (Lusatia), and revealed a similarly high ASD0 value as in the modern Polish population only for R-M17*(xM458) in Kaszuby, which we explained by the presence of an unknown subclade detected in the median-joining network. Apart from R-M17*(xM458) in Kaszuby, genetic diversity for both R-M17 subclades was lower (in several cases much lower) in the native pre-WWII populations than in the modern one. This may be owing to the extensive mixing of the Polish population after the post-WWII massive resettlements, with millions of modern Poles tracing their pre-WWII origin to the Dniester, Dnieper and Neman basins in present-day Ukraine, Belarus and Lithuania.
Kayser et al3 revealed significant genetic differentiation between paternal lineages of neighbouring Poland and Germany, which follows a present-day political border and was attributed to massive population movements during and shortly after the WWII. Although the very recent origin of the geographic course of the detected genetic boundary is undoubted, it remained unknown whether Y-chromosomal diversity in ethnically/linguistically defined Slavic and German populations, which used to be exposed to intensive interethnic contacts and cohabit ethnically mixed territories, was clinal or discontinuous already before the war. In contrast to the regions of Kaszuby and Kociewie, which were politically subordinated to German states for more than three centuries and before the massive human resettlements in the mid-20th century occupied a narrow strip of land between German-speaking territories, the Kurpie region practically never experienced longer periods of German political influence and direct neighbourhood with the German populations. Lusatia was conquered by Germans in the 10th century and since then was a part of German states for most of its history; the modern Lusatians (Sorbs) inhabit a Slavic-speaking island in southeastern Germany. In spite of the fact that these four regions differed significantly in exposure to gene flow with the German population, our results revealed their similar genetic differentiation from Bavaria and Mecklenburg. Moreover, admixture estimates showed hardly detectable German paternal ancestry in Slavs neighbouring German populations for centuries, that is, the Sorbs and Kashubes. However, it should be noted that our regional population samples comprised only individuals of Polish and Sorbian ethnicity and did not involve a pre-WWII German minority of Kaszuby and Kociewie, which owing to forced resettlements in the mid-20th century ceased to exist, and also did not involve Germans constituting since the 19th century a majority ethnic group of Lusatia. Thus, our results concern ethnically/linguistically rather than geographically defined populations and clearly contrast the broad-scale pattern of Y-chromosomal diversity in Europe, which was shown to be strongly driven by geographic proximity rather than by language.1 They are also consistent with a previous study on autosomal markers, which provided evidence for clear genetic departure of the Sorbs from the neighbouring Germans and their genetic similarity to the Slavic-speaking Poles and Czechs.43 Although data for German-speaking populations that used to live in the neighbourhood of the Slavs of Kaszuby and Kociewie are not available, data from the Sorbs and neighbouring Germans could be used as a proxy, and our AMOVA results and ancestry estimates suggest that a genetic barrier between Slavic and German speakers similar to the one detected by Kayser et al3 between modern Poland and Germany might have existed already before the war.
Immel et al44 revealed German and Slavic surname-associated strata in the Halle region in southeastern Germany, which was explained by the 19th century migration from the Polish-speaking territories. As German surnames are frequently encountered among the modern Poles, we have searched for such differentiation within the Polish pre-WWII regional populations. Both Slavic and German surname carriers revealed regional Y chromosome homogeneity and comparable genetic distances from the German populations, which suggests that etymologically German surnames in the studied populations may result, at least partially, from foreign administration and linguistic adaptation (eg, translation, common until the end of the 19th century and attested also in the 20th century), well documented in historical sources,26, 27 rather than owing to genetic admixture.
Two main factors are believed to be responsible for the Slavic language extinction in vast territories to the east of the Elbe and Saale rivers: colonisation of the region by the German-speaking settlers, known in historical sources as Ostsiedlung, and assimilation of the local Slavic populations, but contribution of both factors to the formation of a modern eastern German population used to remain highly speculative.8 Previous studies on Y-chromosomal diversity in Germany by Roewer et al17 and Kayser et al3 revealed east–west regional differentiation within the country with eastern German populations clustering between western German and Slavic populations but clearly separated from the latter, which suggested only minor Slavic paternal contribution to the modern eastern Germans. Our ancestry estimates for the Mecklenburg region (Supplementary Table S3) and for the pooled eastern German populations, assessed as being well below 50%, definitely confirm the German colonisation with replacement of autochthonous populations as the main reason for extinction of local Slavic vernaculars. The presented results suggest that early medieval Slavic westward migrations and late medieval and subsequent German eastward migrations, which outnumbered and largely replaced previous populations, as well as very limited male genetic admixture to the neighbouring Slavs (Supplementary Table S4), were likely responsible for the pre-WWII genetic differentiation between Slavic- and German-speaking populations. Woźniak et al18 compared several Slavic populations and did not detect such a sharp genetic boundary in case of Czech and Slovak males with genetically intermediate position between other Slavic and German populations, which was explained by early medieval interactions between Slavic and Germanic tribes on the southern side of the Carpathians. Anyway, paternal lineages from our Slovak population sample were genetically much closer to their Slavic than German counterparts.
Coalescence-based analysis of populations sharing common ancestry, which experienced subsequent cross-migration, leads to underestimation of their divergence time. On the other hand, coalescence-based analysis of populations sharing common ancestry, which experienced subsequent gene flow with unrelated populations, is likely to overestimate their divergence time and affect other demographic parameters. As the model implemented in BATWING does not assume migration between diverged populations, our analysis was performed on populations of Kaszuby and Lusatia, which owing to geographic remoteness and a linguistic barrier remained isolated from each other and from their German-speaking neighbours. Our coalescence-based divergence time estimates for the two isolated western Slavic populations almost perfectly match historical and archaeological data on the Slavs' expansion in Europe in the 5th–6th centuries.4 Several hundred years of demographic expansion before the divergence, as detected by the BATWING, support hypothesis that the early medieval Slavic expansion in Europe was a demographic event rather than solely a linguistic spread of the Slavic language.
Acknowledgments
We thank all the donors, who have voluntarily donated their biological samples for the study, Professor Thomas Meitinger (Institute of Human Genetics, Neuherberg) for sharing samples, Professor Manfred Kayser (Erasmus University Medical Centre, Rotterdam) for Y-chromosomal lineage data from German regional populations, and Isabel Mendizabal (Institut de Biologia Evolutiva, CSIC-UPF) and Dr David F Soria-Hernanz (National Geographic Society) for an R script implemented in analysis of BATWING output files. BATWING and R calculations were carried out at the Academic Computer Centre in Gdańsk and on a computer cluster at the IBE, CSIC-UPF in Barcelona. KR was supported by the Foundation for Polish Science within the KOLUMB Programme and by the ‘Crescendum Est – Polonia' Foundation. BM-C and DC were supported by MCINN grant CGL2010-14944/BOS.
Appendix
The Genographic Consortium
Syama Adhikarla1, Christina J Adler2, Elena Balanovska3, Oleg Balanovsky3, Jaume Bertranpetit4, Andrew C Clarke5, Alan Cooper2, Clio SI Der Sarkissian2, Matthew C Dulik6, Jill B Gaieski6, ArunKumar GaneshPrasad1, Wolfgang Haak2, Marc Haber4,7, Angela Hobbs8, Asif Javed9, Li Jin10, Matthew E Kaplan11, Shilin Li10, Elizabeth A Matisoo-Smith5, Marta Melé4, Nirav C Merchant11, R John Mitchell12, Amanda C Owings6, Laxmi Parida9, Ramasamy Pitchappan1, Daniel E Platt9, Colin Renfrew13, Daniela R Lacerda14, Ajay K Royyuru9, Fabrício R Santos14, Theodore G Schurr6, Himla Soodyall8, David F Soria Hernanz15, Pandikumar Swamikrishnan16, Chris Tyler-Smith17, Arun Varatharajan Santhakumari1, Pedro Paulo Vieira18, Miguel G Vilar6, R Spencer Wells15, Pierre A Zalloua7, Janet S Ziegle19
Affiliations for participants: 1Madurai Kamaraj University, Madurai, Tamil Nadu, India; 2University of Adelaide, South Australia, Australia; 3Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow, Russia; 4Universitat Pompeu Fabra, Barcelona, Spain; 5University of Otago, Dunedin, New Zealand; 6University of Pennsylvania, Philadelphia, PA, USA; 7Lebanese American University, Chouran, Beirut, Lebanon; 8National Health Laboratory Service, Johannesburg, South Africa; 9IBM, Yorktown Heights, NY, USA; 10Fudan University, Shanghai, China; 11University of Arizona, Tucson, AZ, USA; 12La Trobe University, Melbourne, Victoria, Australia; 13University of Cambridge, Cambridge, UK; 14Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil; 15National Geographic Society, Washington, DC, USA; 16IBM, Somers, NY, USA; 17The Wellcome Trust Sanger Institute, Hinxton, UK; 18Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; 19Applied Biosystems, Foster City, CA, USA
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Rosser ZH, Zerjal T, Hurles ME, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploski R, Wozniak M, Pawlowski R, et al. Homogeneity and distinctiveness of Polish paternal lineages revealed by Y chromosome microsatellite haplotype analysis. Hum Genet. 2002;110:592–600. doi: 10.1007/s00439-002-0728-0. [DOI] [PubMed] [Google Scholar]
- Kayser M, Lao O, Anslinger K, et al. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum Genet. 2005;117:428–443. doi: 10.1007/s00439-005-1333-9. [DOI] [PubMed] [Google Scholar]
- Schenker AM. The Dawn of Slavic: An Introduction to Slavic Philology. New Haven: Yale University Press; 1995. [Google Scholar]
- Norberg M.Die Sorben: slawisches Volk im Osten Deutschlandsin Hinderling R, Eichinger LM, (eds):: Handbuch der mitteleuropäischen Sprachminderheiten Tübingen: Gunter Narr; 1996 [Google Scholar]
- Główny Urząd Statystyczny . Raport z wyników Narodowego Spisu Powszechnego Ludności i Mieszkań 2002. Warszawa: GUS; 2003. [Google Scholar]
- Latoszek M.(ed):: Kaszubi: monografia socjologiczna Rzeszów: Towarzystwo Naukowe Organizacji i Kierownictwa; 1990 [Google Scholar]
- Zaroff R. Germanisation of the land between the Elbe-Saale and the Oder rivers: colonisation or assimilation. Proc Univ Qld Hist Res Group. 1998;9:1–19. [Google Scholar]
- Karaś H.(ed): Dialekty i gwary polskie: kompendium internetowe Zakład Historii Języka Polskiego i Dialektologii Uniwersytetu Warszawskiego & Towarzystwo Kultury Języka; 2010 , http://www.dialektologia.uw.edu.pl/ . [Google Scholar]
- Curta F.From Kossina to Bromley: ethnogenesis in Slavic archaeologyin Gillett A, (ed):: On Barbarian Identity: Critical Approaches to Ethnicity in the Early Middle Ages Turnhout: Brepols; 2002201–218. [Google Scholar]
- Rębała K, Mikulich AI, Tsybovsky IS, et al. Y-STR variation among Slavs: evidence for the Slavic homeland in the middle Dnieper basin. J Hum Genet. 2007;52:406–414. doi: 10.1007/s10038-007-0125-6. [DOI] [PubMed] [Google Scholar]
- Nichols J.The linguistic geography of the Slavic expansionin Maguire RA, Timberlake A, (eds): American Contributions to the Eleventh International Congress of Slavists Columbus: Slavica Publishers; 1993377–391. [Google Scholar]
- Martínez-Cruz B, Harmant C, Platt DE, et al. Evidence of pre-Roman tribal genetic structure in Basques from uniparentally inherited markers. Mol Biol Evol. 2012;29:2211–2222. doi: 10.1093/molbev/mss091. [DOI] [PubMed] [Google Scholar]
- Martínez-Cruz B, Ziegle J, Sanz P, et al. Multiplex single-nucleotide polymorphism typing of the human Y chromosome using TaqMan probes. Investig Genet. 2011;2:13. doi: 10.1186/2041-2223-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Woźniak M, Grzybowski T, Starzyński J, Marciniak T. Continuity of Y chromosome haplotypes in the population of Southern Poland before and after the Second World War. Forensic Sci Int Genet. 2007;1:134–140. doi: 10.1016/j.fsigen.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Roewer L, Croucher PJP, Willuweit S, et al. Signature of recent historical events in the European Y-chromosomal STR haplotype distribution. Hum Genet. 2005;116:279–291. doi: 10.1007/s00439-004-1201-z. [DOI] [PubMed] [Google Scholar]
- Woźniak M, Malyarchuk B, Derenko M, et al. Similarities and distinctions in Y chromosome gene pool of Western Slavs. Am J Phys Anthropol. 2010;142:540–548. doi: 10.1002/ajpa.21253. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Polzin T, Daneshmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20. [Google Scholar]
- Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA. Y-chromosomal evidence for the origins of Oceanic-speaking peoples. Genetics. 2002;160:289–303. doi: 10.1093/genetics/160.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Krawczak M, Excoffier L, et al. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet. 2001;68:990–1018. doi: 10.1086/319510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreja B. Księga nazwisk ziemi gdańskiej. Gdańsk: Wydawnictwo Uniwersytetu Gdańskiego; 1998. [Google Scholar]
- Rymut K. Nazwiska Polaków: słownik historyczno-etymologiczny. Kraków: Wydawnictwo Instytutu Języka Polskiego PAN; 1999–2001. [Google Scholar]
- Breza E. Nazwiska Pomorzan: pochodzenie i zmiany. Gdańsk: Wydawnictwo Uniwersytetu Gdańskiego; 2000–2004. [Google Scholar]
- Wilson IJ, Weale ME, Balding DJ. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J R Stat Soc Ser A Stat Soc. 2003;166:155–201. [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, et al. A predominantly Neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willuweit S, Roewer L. Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007;1:83–87. doi: 10.1016/j.fsigen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Park SW, Hwang CH, Cho EM, Park JH, Choi BO, Chung KW. Development of a Y-STR 12-plex PCR system and haplotype analysis in a Korean population. J Genet. 2009;88:353–358. doi: 10.1007/s12041-009-0053-7. [DOI] [PubMed] [Google Scholar]
- Weale ME, Weiss DA, Jager RF, Bradman N, Thomas MG. Y chromosome evidence for Anglo-Saxon mass migration. Mol Biol Evol. 2002;19:1008–1021. doi: 10.1093/oxfordjournals.molbev.a004160. [DOI] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Bertorelle G. Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol. 2001;18:672–675. doi: 10.1093/oxfordjournals.molbev.a003847. [DOI] [PubMed] [Google Scholar]
- Wang J. Maximum-likelihood estimation of admixture proportions from genetic data. Genetics. 2003;164:747–765. doi: 10.1093/genetics/164.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Nicholson J, et al. Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet. 2000;67:697–717. doi: 10.1086/303046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Myres NM, Rootsi S, et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet. 2010;18:479–484. doi: 10.1038/ejhg.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myres NM, Rootsi S, Lin AA, et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Główny Urząd Statystyczny . Narodowy Spis Powszechny z dnia 3 grudnia 1950 r.: miejsce zamieszkania ludności w sierpniu 1939 r. Warszawa: GUS; 1955. [Google Scholar]
- Veeramah KR, Tönjes A, Kovacs P, et al. Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity. Eur J Hum Genet. 2011;19:995–1001. doi: 10.1038/ejhg.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immel U-D, Krawczak M, Udolph J, et al. Y-chromosomal STR haplotype analysis reveals surname-associated strata in the East-German population. Eur J Hum Genet. 2006;14:577–582. doi: 10.1038/sj.ejhg.5201572. [DOI] [PubMed] [Google Scholar]
- Balaresque P, Bowden GR, Parkin EJ, et al. Dynamic nature of the proximal AZFc region of the human Y chromosome: multiple independent deletion and duplication events revealed by microsatellite analysis. Hum Mutat. 2008;29:1171–1180. doi: 10.1002/humu.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.