Abstract
Objective
Obesity has become a worldwide health problem in the past decades. Human and animal studies have implicated serotonin in appetite regulation, and behavior genetic studies have shown that body mass index (BMI) has a strong genetic component. However, the roles of genes related to the serotoninergic (5-hydroxytryptamine,5-HT) system in obesity/BMI are not well understood, especially in Chinese subjects.
Subjects and Design
With a sample of 478 healthy Chinese volunteers, this study investigated the relation between BMI and genetic variations of the serotoninergic system as characterized by 136 representative polymorphisms. We used a system-level approach to identify SNPs associated with BMI, then estimated their overall contribution to BMI by multiple regression and verified it by permutation.
Results
We identified 12 SNPs that made statistically significant contributions to BMI. After controlling for gender and age, four of these SNPs accounted for 7.7% additional variance of BMI. Permutation analysis showed that the probability of obtaining these findings by chance was low (p = 0.015, permuted for 1000 times).
Conclusion
These results showed that genetic variations in the serotoninergic system made a moderate contribution to individual differences in BMI among a healthy Chinese sample, suggesting that a similar approach can be used to study obesity.
Introduction
A decade ago, the World Health Organization warned about a growing obesity epidemic and listed more than 30 diseases that are causally related to obesity [1]. Globally, approximately 1.6 billion adults are either overweight (BMI [weight in kilogram divided by the square of height in meter]≥25) or obese (BMI ≥30) [2]. In fact, the rates of obesity have tripled in developing countries in the past 20 years [3]. Moreover, childhood obesity is also increasing rapidly worldwide [4].
Although many environmental factors (e.g. freely available high-calorie food, sedentary life style, low socio-economic status and high-danger neighborhood environment) predispose individuals to gaining weight [5], [6], [7], [8], genetic factors also contribute to energy homeostasis or appetite, which can lead to obesity. Family, twin, and adoption studies indicate that 24%–90% of human BMI variation is due to genetic factors [9], [10], [11], [12], [13], [14]. Recent molecular genetic studies have identified many genes that regulate appetite or energy balance (e.g, FTO, MC4R, SH2B1, and serotonin related genes ) and have robust associations with obesity or BMI [15], [16].
Because serotonin can regulate appetite by activating pro-opiomelanocortin (POMC) neurons, which play a key role in the regulation of feeding by sending anorectic signals to the periventricular nucleus (PVN) and other brain areas associated with energy homeostasis [17], serotonin as well as related genes are often tested for association with weight gain and obesity. Indeed, a strong negative correlation between blood 5-HT concentration and body mass was found both in mice [18] and in human [19]. Studies of SERT knockout mice have uncovered SERT as a candidate gene for obesity, with SERT mutant (SCL6A4−/−) mice becoming obese [20]. This polymorphism has also been associated in some studies with eating disorder [21], [22], [23] and obesity [24], [25], although other studies showed no association between the 5-HTTLPR polymorphism and weight regulation [26], [27], [28]. In terms of the 5-HT receptor genes, the serotonin (5-HT) receptor HTR2C was demonstrated to play a role in modulating appetite behavior using knock-out mice [29], [30], normal population [31] and patients [32], [33], [34], [35], although some studies [36], [37] failed to replicate that result. HTR1B [38], [39], HTR2A [40], [41], HTR3B [42] were also reported to be associated with body mass or obesity. MAOA was also found to influence body mass [43] or obesity [44].
Although these serotonin-related genes have been identified as being relevant to body mass and obesity, the results have not always been consistent and the size of their effects has been typically small, far less than previously estimated 24%–90% heritability. There may be many reasons for these inconsistent results and small effect sizes. One most likely reason is polygenicity. Complex quantitative traits are influenced by many genes, each with a small effect. As early as 1918, Fisher proposed this polygenic model that combined many genes of small effects to yield the continuous variation for most quantitative traits [45]. Recently, some studies have successfully applied the polygenic model by combining effects of the whole genome [46], [47], [48] or effects of genes within a pathway [49], [50], [51]. Since several serotonin-related genes exert their effect on BMI, it is likely that their effects are cumulative. The current study used a system-level approach to examine the role of the serotoninergic system in BMI/obesity.
Another possible reason for inconsistent results may be the heterogeneity in samples across studies. Subjects in different studies differ in their health status, age, sex, and ethnicity, which might have confounded the relations between genes and BMI. For example, associations between 5HTR2A and BMI are found in obese [40] and anorexia nervosa patients [41] but not in healthy controls. Similarly, 5HTTLPR was associated with BMI in non-elderly (<65 yr) stroke patients but not in elderly patients (> or = 65 yr). An association was observed between MAOA and obesity among white and Hispanic American subjects, but not among African–American subjects [44]. Thus it is important to control for these potential confounding factors.
The current study adopted the system-level approach to examine the role of the serotoninergic system in body mass in a relatively homogenous sample (in terms of age, health status, and ethnicity). We enrolled a sample of young healthy Han Chinese subjects, genotyped polymorphisms within the serotonin system, and calculated their BMI. Specifically, we selected 136 polymorphic loci (including 134 SNPs and 2 VNTR polymorphisms) to cover a substantial portion (by LD) of the common variations within known genes of the 5-HT system to estimate the additive and multiplicative contributions of these genes on BMI.
Materials and Methods
Participants
Four hundred and eighty healthy Chinese college students (mean age = 20, SD = 1) were recruited from Beijing Normal University, Beijing, China. They had normal or corrected-to-normal vision, and had no history of neurological or psychiatric problems according to self-report. None of them were identified as having alcohol or nicotine dependence according to the Alcohol Use Disorders Identification Test [52] and the Fagerström Test for Nicotine Dependence [53]. Two participants were excluded because of poor genotyping results. A written consent form was obtained from each subject after a full explanation of the study procedure. This study was approved by the IRB of the State Key Laboratory of Cognitive Neuroscience and Learning at Beijing Normal University, China.
BMI Measurements
Height and weight of subjects were self-reported. BMI was calculated as weight (kg) divided by the square of height (m). Self-reported data on weight and height have been used by previous large-scale studies on body mass and proved to be highly reliable in calculating BMI [22], [38], [46], [54], [55], [56], [57]. Furthermore, all students including all of our participants were given an annual physical examination at the beginning of the academic year in September and they were informed of their height and weight. Self-report data on height and weight were collected in December.
Genetic Analysis
Gene selection
We selected 25 genes and 136 associated polymorphisms (134 SNPs and 2 VNTR polymorphisms) distributed across the synthesis, degradation, transporter, and receptor subsystems of the 5-HT system. 5-HT synthesis involves converting the tryptophan (via TPH) to 5-hydroxytryptophan (5-HTP), followed by subsequent hydroxylation (by TPH) to 5-HT. We included two genes related to 5-HT synthesis: tryptophan hydroxylase (TPH1 and TPH2, with three SNPs each). For the degradation subsystem, released 5-HT is directly broken down at the synapse into inactive metabolites by two enzymes, COMT and MAO (including MAOA and MAOB). We included catechol-O-methyl transferase gene (COMT, with 7 SNPs) and monoamine oxidase genes (MAOA, with 5 SNPs and 1 VNTR, and MAOB with 3 SNPs). The 5-HT transporter includes (1) SLC6A4, an integral membrane-spanning protein that pumps the neurotransmitter serotonin from synaptic spaces into presynaptic neurons and (2) VMAT, a transport protein integrated into the membrane of intracellular vesicles of presynaptic neurons, which acts to transport monoamines into the synaptic vesicles. We included SLC6A4 (7 SNPs plus 5HTTLPR), VMAT1 (SLC18A1, 9 SNPs), and VMAT2 (SLC18A2, 5 SNPs). For the receptor subsystem, we included all 17 genes (with the respective number of SNPs in parentheses): HTR1A (2), HTR1B (2), HTR1D (13), HTR1F (5), HTR2A (21), HTR2B (6), HTR2C (3), HTR3A (1), HTR3B (2), HTR3C (3), HTR3D (4), HTR3E (2), HTR4 (10), HTR5A (4), HTR5B (2), HTR6 (5), and HTR7 (7). Together, the above 25 genes represent all major genes involved in these four 5-HT subsystems in humans [58]. Details about these genes and the selected loci can be found in Table S1.
Genotyping techniques
The SNPs were genotyped using the standard Illumina Golden Gate Genotyping protocol (see Illumina Golden-Gate Assay Protocol for details, http://www.southgene.com.cn; Shanghai South Gene Technology Co., Ltd, Shanghai, China). In addition, three genetic markers (5HTTLPR, MAOA VNTR, and COMT rs4680) were ascertained by standard PCR procedures [59], [60], [61].
Gene data preprocessing
Two subjects with more than 10% null genotyping were excluded. In addition to automatic calling of genotypes, Illumina genotyping platform supplied a quantitative quality measure known as the GenCall score. It measures how close a genotype is to the center of the cluster of other samples assigned to the same genotypes, compared with the centers of the clusters of the other genotypes. This measure ranges from 0 to 1, with a higher score indicating a more reliable result. The conventional cutoff point is.25 [62]. Of the 63574 genotypes (133 SNPs by 478 subjects) in the current study, 120 genotypes (0.2%) were excluded because their GenCall scores were lower than.25.
Additional data cleaning included the treatment of low-frequency alleles. For SNPs with either heterozygote or minor homozygote found in fewer than 10 (about 2%) participants, these two genotype groups were combined. If the combined group still had fewer than 10 participants, the SNP(s) were excluded from further analysis. SNPs that showed no polymorphisms were also deleted. In order to examine sample representativeness, Hardy-Weinberg equilibrium (HWE) index was calculated using the Chi square test and setting df to 1. Since males have only one X chromosome, only females were included in HWE calculation for SNPs located on X chromosome. Five of the SNPs showed significant HW disequilibrium (p<0.01). The inclusion of both tSNPs and additional SNPs in regions detected in selection screens [63], [64] resulted in high LD among a number of SNPs. Thirty-one SNPs included in initial analysis were excluded from multiple regression analysis because of their high LD with other adjacent SNPs (R2>0.8, calculated with Plink [65]), yielding a final list of 105 polymorphisms for the main data analyses. It is worth mentioning that the “redundant” SNPs showed the same or almost the same results as the linked SNPs, confirming the association. Table S1 shows the details about all 136 polymorphic loci (134 SNPs and 2 VNTRs) included in our study: location (rs number, chromosome, position), gene, serotonin subsystem, allele polymorphism and frequency, Hardy Weinberg equilibrium, linkage disequilibrium and deleted SNPs. Finally, genetic relatedness of subjects was checked following Anderson et.al. [66] protocol using Plink. We used all 240 unrelated autosome SNPs (r2<0.8) available in the larger project of these subjects and set the threshold of 0.95 (personal communication with Drs. Anderson and Zondervan). We found no pair of subjects showing high relatedness (all PI_HAT smaller than or equal to 0.5).
Data Analysis
The goal of the current study was to understand the relation between individual differences in BMI and genetic variations in the 5-HT system in healthy subjects. Moving beyond the single-gene or a small number of haplotypes approaches used in typical molecular behavior genetics research, this study used the system-level approach [50] to examine the overall contributions of the serotoninergic system (characterized by the major genes and their associated loci) on BMI.
Briefly, the analysis includes three steps: First, ANOVA was used to screen polymorphism loci that showed nominal significance (p<0.05) on BMI; these loci were then entered into a regression model to estimate their overall contribution to BMI after controlling for gender and age; and lastly the regression model was verified by permutation. In this study, we built two kinds of regression models. In model 1 (main effects), we included the loci with significant main effects based on the ANOVA results and used the forward stepwise method to build the model. Gender and age were entered as control variables. To run multiple regression analyses, all SNPs were coded in a linear way, i.e. the major homozygote, heterozygote, and minor homozygote were coded into 1, 2, and 3, respectively (SNPs on X chromosome were coded as 1 and 3 for major and minor allele, and 3 for female heterozygotes). In addition, the MAOA VNTR was coded as 1 for the 3 repeat and 3 for the 4 repeat in males and 1 for 3 repeat homozygotes and 3 for others in females. In model 2, all two-way interactions of these loci in model 1 were added using forward stepwise method. Permutation was done 1000 times by shuffling BMI (along with gender and age) across subjects, and the probability of getting a larger R2 in the shuffled data than in the real data was defined as p value of the model.
Results
The mean BMI for our sample was 20.5 kg/m2 (SD = 2.4), ranging from 16.3 to 37.5. According to WHO BMI classification, there were 93 (71 female) underweight participants (BMI <18.5), 359 (192 female) normal weight participants (18.5≤ BMI <25), and 26 (8 female) overweight participants (BMI ≥25). The BMI distribution in the present study was comparable to other studies with Chinese college students [67], [68]. Males (21.14±2.44) had significantly higher BMI than females (20.00±2.67; t(476) = 5.30, p = 1.8E−7), which was consistent with previous findings in healthy young Chinese [69].
Of the 105 SNPs, 12 showed significant main effects with uncorrected p<0.05. Specifically, individuals with the following genotypes showed lower BMI than those with alternative alleles: homozygous for the major allele of rs13166761 (HTR4), rs1018079 (SLC18A1(VMAT1)), rs11214769 (HTR3B), rs977003 (HTR2A), rs2224721 (HTR2A), rs2192371 (HTR2C), rs4911871 (HTR2C), or rs2270638 (VMAT1); or heterozygous/minor allele homozygous for rs6651806 (MAOB), rs5905512 (MAOB); or homozygous for the minor allele of rs7904569 (HTR7) or rs6644065 (HTR2C) (see Table 1, and Table S2 for effects of all loci). These SNPs were used in a regression analysis to build model 1 (main effects). There was no significant gender-by-SNP interaction except rs5905512 (see Table 1), and this SNP did not contribute to regression model 1, so we included gender, but not gender-by-gene interactions, as a covariate in the following analysis.
Table 1. Means and standard deviations of BMI for each polymorphism, and main effects and post hoc comparisons of SNPs that showed significant main effects and were used in subsequent multiple regression analysis.
| SNP | Subsystem | Gene | Maj | Mean | SD | n | Het | Mean | SD | n | Min | Mean | SD | n | F | p | mh | mm | hma | Gene bygenderinteractionF(p) |
| rs6651806 | Degradation | MAOB | AA | 20.63 | 2.45 | 381 | AC | 19.95 | 2.16 | 97 | CC | 6.32 | 0.01 | b | 0.40 (0.53) | |||||
| rs5905512 | MAOB | AA | 20.76 | 2.62 | 284 | AG | 20.10 | 2.01 | 194 | GG | 9.02 | <0.01 | b | 0.02 (0.89) | ||||||
| rs1018079 | Transport | SLC18A1 | AA | 20.29 | 2.26 | 303 | AG | 20.72 | 2.32 | 156 | GG | 22.10 | 4.35 | 18 | 5.92 | <0.01 | 0.07 | <0.01 | 0.02 | 0.34 (0.71) |
| rs2270638 | SLC18A1 | AA | 20.33 | 2.47 | 344 | AG | 20.92 | 2.20 | 133 | GG | 5.77 | 0.02 | b | 0.27 (0.60) | ||||||
| rs977003 | Receptor | HTR2A | AA | 20.29 | 2.30 | 299 | AC | 20.92 | 2.62 | 151 | CC | 20.40 | 2.19 | 28 | 3.45 | 0.03 | 0.01 | 0.82 | 0.29 | 0.14 (0.87) |
| rs2224721 | HTR2A | CC | 20.12 | 2.12 | 216 | AC | 20.79 | 2.61 | 209 | AA | 20.87 | 2.51 | 53 | 4.85 | 0.01 | <0.01 | 0.04 | 0.82 | 0.13 (0.88) | |
| rs2192371 | HTR2C | AA | 20.61 | 2.42 | 242 | AG | 19.79 | 1.80 | 124 | GG | 21.03 | 2.78 | 112 | 8.62 | <0.01 | <0.01 | 0.12 | <0.01 | 0.95 (0.33) | |
| rs6644065 | HTR2C | AA | 20.45 | 2.26 | 373 | AG | 20.05 | 2.81 | 68 | GG | 21.74 | 2.77 | 37 | 6.30 | <0.01 | 0.21 | <0.01 | <0.01 | 6.06 (0.01) | |
| rs4911871 | HTR2C | AA | 20.39 | 2.23 | 350 | AG | 20.17 | 2.74 | 76 | GG | 21.68 | 2.76 | 51 | 7.44 | <0.01 | 0.47 | <0.01 | <0.01 | 1.09 (0.30) | |
| rs11214769 | HTR3B | AA | 20.31 | 2.22 | 335 | AG | 20.88 | 2.85 | 124 | GG | 21.21 | 2.20 | 19 | 3.46 | 0.03 | 0.02 | 0.11 | 0.58 | 1.65 (0.19) | |
| rs13166761 | HTR4 | GG | 20.38 | 2.23 | 244 | AG | 20.79 | 2.64 | 202 | AA | 19.48 | 1.81 | 32 | 4.74 | 0.01 | 0.07 | 0.05 | <0.01 | 0.54 (0.59) | |
| rs7904569 | HTR7 | AA | 20.50 | 2.38 | 206 | AG | 20.69 | 2.53 | 215 | GG | 19.72 | 1.85 | 57 | 3.64 | 0.03 | 0.43 | 0.03 | 0.01 | 1.34 (0.26) |
Note: Empty cells mean no such genotypes were found in our sample. Maj: Major allele; Het: Heterozygote; Min: Minor allele.
Results (p values) of post hoc comparisons. mh = Maj versus Het, mm = Maj versus Min, hm = Het versus Min.
Post hoc comparison was not run because there were only 2 groups for this locus.
Table 2 shows the results of the multiple regression analysis. On the first step, two control variables (gender and age) were entered. Together they accounted for 5.6% variance of BMI. On the second step, forward stepwise regression resulted in four of the 12 SNPs to be included in the regression equation, showing that they made unique contributions to explaining variance in BMI. Together these SNPs accounted for 7.7% additional variance, yielding a total R2 of.13, F(6,455) = 11.61, p = 4.08E−12.
Table 2. Regression models.
| Regressor | Gene | Beta | T | P |
| Gender | −0.24 | −5.39 | 0.00 | |
| Age | 0.02 | 0.47 | 0.64 | |
| rs1018079 | SLC18A1(VMAT1) | 0.16 | 3.63 | 0.00 |
| rs11214769 | HTR3B | 0.14 | 3.08 | 0.00 |
| rs2224721 | HTR2A | 0.12 | 2.76 | 0.01 |
| rs4911871 | HTR2C | 0.12 | 2.81 | 0.01 |
Note: ‘Gene’ is the corresponding gene for each SNP; ‘beta’ is the standardized regression coefficient, ‘T’ and ‘P’ are t-test results.
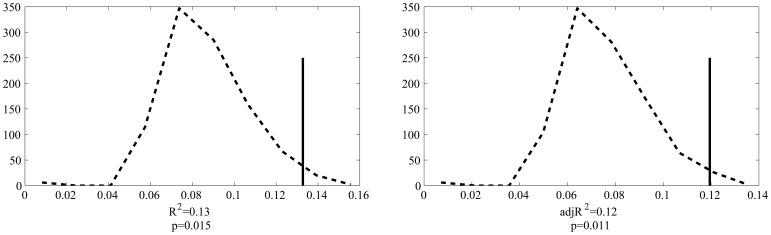
Permutation results are shown in Figure 1. Based on 1000 permutations, the probability of attaining the R2 or adjusted R2 found in our model was 0.015 and 0.011, respectively.
Figure 1. Permutation results for R2 (left panel) and adjusted R2 (right panel).
Dashed line represents distribution of R2 obtained from randomized data and solid line represents the observed R2.
We then added potential interactive effects to investigate whether additional variance in BMI can be accounted for by gene–gene interactions. In this analysis, we first entered the control variables (gender and age) and the four SNPs in model 1 and finally their two-way interactions using the stepwise procedure. For the four SNPs that entered model 1, there were 6 potential interactions. None of the interaction terms made significant and unique contributions to the model.
Discussion
Based on the system-level analysis of 5-HT neurotransmitter genes, we identified 12 SNPs of the 5-HT-related genes showing nominal effects on BMI. Four of these SNPs made significant unique contributions to BMI even after controlling for gender and age. This result has two significant implications. First, the current study revealed a significant role for genes in the 5-HT system on BMI among Chinese, confirming that body mass is likely to be influenced to some extent by the serotoninergic system. Second, our results supported the idea that BMI may be determined by many loci. Only by summing up their overall effects can we understand the genetic basis of a complex trait such as body mass. This approach can estimate the overall contribution of genes within a pathway and can help to explain the missing heritability [46].
We found that 12 SNPs of seven genes (MAOB, SLC18A1(VMAT1), HTR2A, HTR2C, HTR3B, HTR4, and HTR7) were significantly associated with BMI. As summarized in the introduction, previous studies have already found evidence, although not always consistent, of association between the HTR2A and HTR2C genes and BMI. However, other genes we identified have not been tested previously in BMI-related studies to the best of our knowledge.
VMAT1 is expressed primarily in neuroendocrine cells such as the adrenal medulla and pineal gland [70], [71], [72]. As early as in 1999, Hayashi et al. [73] found that VMAT1 was responsible for the storage of 5-hydroxytryptamine in rat pinealocytes. Mammalian pinealocytes contain more 5-HT than any other cells. Upon stimulation by norepinephrine (NE), the internal 5-HT is released and then stimulates serotonin N-acetyltransferase activity via the 5-HT2 receptor, resulting in increased melatonin output [73]. Melatonin has been found to be involved in energy metabolism and body weight control in both animals [74], [75] and humans [76]. Decreased activity of the melanocortin system produces a marked orexigenic effect, while increased activity increases α-melanocyte-stimulating hormone (α-MSH) release leading to satiation and a termination of feeding. On the other hand, the missence variation Thr136Ile in the VMAT1/SLC18A1 gene was found to be associated with anxiety-related personality traits [77] and anxiety has been shown to be associated with obesity [78], [79] or BMI [80]. Previous studies have found that VMAT plays an important role in the life cycle of ghrelin and obestatin in the A-like cells of the stomach [81], [82], and ghrelin and obestatin have effects on food intake and energy balance. Therefore, we speculate that the VMAT1 gene may have an effect on BMI through melatonin output, ghrelin, obestatin or anxiety mood. This gene accounted for the largest proportion of the variance of BMI in our study (Table 2).
The 5-HT3 receptor has been suggested to be involved in anxiety, depression, pain, alcohol dependence, and eating disorders [42], [83]. The HTR3B gene encodes the B-subunit of the type 3 serotonin receptor (5-HT3), a ligand-gated ion channel that is known to be involved in gut motility and peristalsis. Thus the HTR3B gene may regulate BMI because gut motility is associated with numerous gastrointestinally derived peptides with significant effects on food intake and energy balance [84]. Many studies have also reported that the 5-HT3B polymorphism is associated with the incidence of major depression [85], efficiency of the antidepressant treatment [86], and the incidence and severity of nausea after paroxetine treatment of psychiatric patients [87]. Although the specific biological mechanisms are not well understood, our results indicate that HTR3B gene polymorphism may influence body mass via gut motility or mood.
Our analysis also showed that HTR2C and HTR2A are possible factors influencing BMI in Chinese subjects, as have been reported by previous studies. Different from the most often studied C759T polymorphism associated with weight gain [31], [32], [33], [34], [35], [88], three SNP we found related to BMI are all located in the intron region of HTR2C. First, there is strong evidence for an interaction between leptin and the 5-HTergic system [88]. Second, McCarthy et al. showed a strong effect of HTR2C polymorphism −759G>A on circulating leptin levels after adjusting for body fat. Other studies also suggested that serotonin influences food intake because of variations in the HTR2C receptor [89], [90]. Similarly, previous researchers have also found an association between a polymorphism −1438G4A (rs6311) in the regulatory region of the HTR2A gene and alteration in food intake [91], [92], [93], [94], but the significant SNP rs2224721 we found is intronic. Recent studies suggest that polymorphic variation in the HTR2A gene may be associated with abdominal obesity and the metabolic syndrome, and that HTR2A may be linked to the stability of the stress-related system (i.e., the serotonin-hypothalamic-pituitary-adrenal system) [95], [96].
Several limitations of the current study need to be mentioned. First, height and weight of the subjects were self-reported. Although other large-scale studies also used self-reported data [22], [38], [46], [54], [55], [56], [57] and previous research showed high correlations (r = .92) between BMI calculated from self-reports and that from actual measurements [56], it would still be better to measure weight and height during the experiment. Second, this study focused only on healthy Han Chinese college students, so these results may or may not be generalized to other populations (e.g., clinical samples, other ethnic groups). Third, the sample size of the current study is modest. As power calculations based on the effect sizes of established variants have suggested that increasing the sample size would likely lead to the discovery of additional variants [97], follow-up research needs to expand the sample size and validate the results. Fourth, we examined only the serotonin system and accounted for only 7% of the variance of BMI, there is much more “missing heritability” of BMI (estimated 24–90%) to be accounted for. Other genetic systems [15], [16] and behavioral factors (e.g, such as physical activity, sedentary life-style, and dietary patterns) as well as their interactions need to be examined. Furthermore, gene-environment interaction studies are needed to understand epigenetic factors in BMI.
In conclusion, we used a system-level approach to identify several genetic SNPs associated with variations in BMI. This analysis provides further evidence for the association between genetic variants in the serotonin pathway and BMI. Because current lifestyle interventions are largely ineffective in addressing the challenges of growing obesity [98], [99], new insights into the biology of obesity are critically needed to guide the development and application of future therapies and interventions.
Supporting Information
Detailed information of the loci used in this study.
(DOC)
Means and standard deviations of BMI for each polymorphism, and main effects and post hoc comparisons of each locus.
(DOC)
Acknowledgments
We thank all graduate research assistants who helped with data collection and the reviewers for their insightful comments.
Funding Statement
This study was supported by the 111 Project (B07008) of the Ministry of Education of China, National Natural Science Foundation of China (31100807), and The Research Fund for the Dectoral Program of Higher Education (20110003120001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2000) Obesity: preventing and managing the global epidemic. Report of a WHO ConsultationWHO Technical Report Series 894Geneva. [PubMed]
- 2.WHO (2009) Global Database on Body Mass Index (BMI). Geneva, Switzerland: World Health Organization.
- 3. Hossain P, Kawar B, El Nahas M (2007) Obesity and diabetes in the developing world–a growing challenge. N Engl J Med 356: 213–215. [DOI] [PubMed] [Google Scholar]
- 4. Lobstein T, Baur L, Uauy R (2004) Obesity in children and young people: a crisis in public health. Obes Rev 5 Suppl 1: 4–104. [DOI] [PubMed] [Google Scholar]
- 5. Bloom SR, Kuhajda FP, Laher I, Pi-Sunyer X, Ronnett GV, et al. (2008) The obesity epidemic: pharmacological challenges. Mol Interv 8: 82–98. [DOI] [PubMed] [Google Scholar]
- 6. Mekhmoukh A, Chapelot D, Bellisle F (2012) Influence of environmental factors on meal intake in overweight and normal-weight male adolescents. A laboratory study. Appetite 59: 90–95. [DOI] [PubMed] [Google Scholar]
- 7. Story M, Sallis JF, Orleans CT (2009) Adolescent obesity: towards evidence-based policy and environmental solutions. J Adolesc Health 45: S1–5. [DOI] [PubMed] [Google Scholar]
- 8. Saelens BE, Sallis JF, Frank LD, Couch SC, Zhou C, et al. (2012) Obesogenic neighborhood environments, child and parent obesity: the Neighborhood Impact on Kids study. Am J Prev Med 42: e57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fabsitz RR, Sholinsky P, Carmelli D (1994) Genetic influences on adult weight gain and maximum body mass index in male twins. Am J Epidemiol 140: 711–720. [DOI] [PubMed] [Google Scholar]
- 10. Austin MA, Friedlander Y, Newman B, Edwards K, Mayer-Davis EJ, et al. (1997) Genetic influences on changes in body mass index: a longitudinal analysis of women twins. Obes Res 5: 326–331. [DOI] [PubMed] [Google Scholar]
- 11. Maes HH, Neale MC, Eaves LJ (1997) Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 27: 325–351. [DOI] [PubMed] [Google Scholar]
- 12. Fox CS, Heard-Costa NL, Vasan RS, Murabito JM, D’Agostino RB Sr, et al. (2005) Genomewide linkage analysis of weight change in the Framingham Heart Study. J Clin Endocrinol Metab 90: 3197–3201. [DOI] [PubMed] [Google Scholar]
- 13. Hjelmborg JB, Fagnani C, Silventoinen K, McGue M, Korkeila M, et al. (2008) Genetic influences on growth traits of BMI: a longitudinal study of adult twins. Obesity (Silver Spring) 16: 847–852. [DOI] [PubMed] [Google Scholar]
- 14. Wardle J, Carnell S, Haworth CM, Plomin R (2008) Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr 87: 398–404. [DOI] [PubMed] [Google Scholar]
- 15. Walley AJ, Asher JE, Froguel P (2009) The genetic contribution to non-syndromic human obesity. Nature Reviews Genetics 10: 431–442. [DOI] [PubMed] [Google Scholar]
- 16. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sargent BJ, Moore NA (2009) New central targets for the treatment of obesity. Br J Clin Pharmacol 68: 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Albay R, Chen A, Anderson GM, Tatevosyan M, Janušonis S (2009) Relationships among body mass, brain size, gut length, and blood tryptophan and serotonin in young wild-type mice. BMC Physiology 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Irish M, Addis DR, Hodges JR, Piguet O (2012) Considering the role of semantic memory in episodic future thinking: evidence from semantic dementia. Brain 135: 2178–2191. [DOI] [PubMed] [Google Scholar]
- 20. Murphy DL, Lesch KP (2008) Targeting the murine serotonin transporter: insights into human neurobiology. Nat Rev Neurosci 9: 85–96. [DOI] [PubMed] [Google Scholar]
- 21. Monteleone P, Santonastaso P, Mauri M, Bellodi L, Erzegovesi S, et al. (2006) Investigation of the serotonin transporter regulatory region polymorphism in bulimia nervosa: relationships to harm avoidance, nutritional parameters, and psychiatric comorbidity. Psychosom Med 68: 99–103. [DOI] [PubMed] [Google Scholar]
- 22. van Strien T, van der Zwaluw CS, Engels RC (2010) Emotional eating in adolescents: a gene (SLC6A4/5-HTT) - depressive feelings interaction analysis. J Psychiatr Res 44: 1035–1042. [DOI] [PubMed] [Google Scholar]
- 23. Calati R, De Ronchi D, Bellini M, Serretti A (2011) The 5-HTTLPR polymorphism and eating disorders: a meta-analysis. Int J Eat Disord 44: 191–199. [DOI] [PubMed] [Google Scholar]
- 24. Lan MY, Chang YY, Chen WH, Kao YF, Lin HS, et al. (2009) Serotonin transporter gene promoter polymorphism is associated with body mass index and obesity in non-elderly stroke patients. J Endocrinol Invest 32: 119–122. [DOI] [PubMed] [Google Scholar]
- 25. Sookoian S, Gianotti TF, Gemma C, Burgueno A, Pirola CJ (2008) Contribution of the functional 5-HTTLPR variant of the SLC6A4 gene to obesity risk in male adults. Obesity (Silver Spring) 16: 488–491. [DOI] [PubMed] [Google Scholar]
- 26. Mergen H, Karaaslan C, Mergen M, Deniz Ozsoy E, Ozata M (2007) LEPR, ADBR3, IRS-1 and 5-HTT genes polymorphisms do not associate with obesity. Endocr J 54: 89–94. [DOI] [PubMed] [Google Scholar]
- 27.Hinney A, Barth N, Ziegler A, von Prittwitz S, Hamann A, et al. (1997) Serotonin transporter gene-linked polymorphic region: allele distributions in relationship to body weight and in anorexia nervosa. Life Sci 61: PL 295–303. [DOI] [PubMed]
- 28.Shinozaki G, Romanowicz M, Kung S, Rundell J, Mrazek D (2012) Investigation of serotonin transporter gene (SLC6A4) by child abuse history interaction with body mass index and diabetes mellitus of White female depressed psychiatric inpatients. Psychiatr Genet. [DOI] [PubMed]
- 29. Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, et al. (1995) Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374: 542–546. [DOI] [PubMed] [Google Scholar]
- 30. Bickerdike MJ, Vickers SP, Dourish CT (1999) 5-HT2C receptor modulation and the treatment of obesity. Diabetes Obes Metab 1: 207–214. [DOI] [PubMed] [Google Scholar]
- 31. Bah J, Westberg L, Baghaei F, Henningsson S, Rosmond R, et al. (2010) Further exploration of the possible influence of polymorphisms in HTR2C and 5HTT on body weight. Metabolism 59: 1156–1163. [DOI] [PubMed] [Google Scholar]
- 32. Godlewska BR, Olajossy-Hilkesberger L, Ciwoniuk M, Olajossy M, Marmurowska-Michalowska H, et al. (2009) Olanzapine-induced weight gain is associated with the −759C/T and −697G/C polymorphisms of the HTR2C gene. Pharmacogenomics J 9: 234–241. [DOI] [PubMed] [Google Scholar]
- 33. Hoekstra PJ, Troost PW, Lahuis BE, Mulder H, Mulder EJ, et al. (2010) Risperidone-induced weight gain in referred children with autism spectrum disorders is associated with a common polymorphism in the 5-hydroxytryptamine 2C receptor gene. J Child Adolesc Psychopharmacol 20: 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryu S, Cho EY, Park T, Oh S, Jang WS, et al. (2007) −759 C/T polymorphism of 5-HT2C receptor gene and early phase weight gain associated with antipsychotic drug treatment. Prog Neuropsychopharmacol Biol Psychiatry 31: 673–677. [DOI] [PubMed] [Google Scholar]
- 35. Gunes A, Melkersson KI, Scordo MG, Dahl ML (2009) Association between HTR2C and HTR2A polymorphisms and metabolic abnormalities in patients treated with olanzapine or clozapine. J Clin Psychopharmacol 29: 65–68. [DOI] [PubMed] [Google Scholar]
- 36. Vimaleswaran KS, Zhao JH, Wainwright NW, Surtees PG, Wareham NJ, et al. (2010) Association between serotonin 5-HT-2C receptor gene (HTR2C) polymorphisms and obesity- and mental health-related phenotypes in a large population-based cohort. Int J Obes (Lond) 34: 1028–1033. [DOI] [PubMed] [Google Scholar]
- 37. Park YM, Cho JH, Kang SG, Choi JE, Lee SH, et al. (2008) Lack of association between the −759C/T polymorphism of the 5-HT2C receptor gene and olanzapine-induced weight gain among Korean schizophrenic patients. J Clin Pharm Ther 33: 55–60. [DOI] [PubMed] [Google Scholar]
- 38. Edwards TL, Velez Edwards DR, Villegas R, Cohen SS, Buchowski MS, et al. (2012) HTR1B, ADIPOR1, PPARGC1A, and CYP19A1 and obesity in a cohort of Caucasians and African Americans: an evaluation of gene-environment interactions and candidate genes. Am J Epidemiol 175: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levitan RD, Kaplan AS, Masellis M, Basile VS, Walker ML, et al. (2001) Polymorphism of the serotonin 5-HT1B receptor gene (HTR1B) associated with minimum lifetime body mass index in women with bulimia nervosa. Biol Psychiatry 50: 640–643. [DOI] [PubMed] [Google Scholar]
- 40. Sorli JV, Frances F, Gonzalez JI, Guillen M, Portoles O, et al. (2008) Impact of the −1438G>a polymorphism in the serotonin 2A receptor gene on anthropometric profile and obesity risk: a case-control study in a Spanish Mediterranean population. Appetite 50: 260–265. [DOI] [PubMed] [Google Scholar]
- 41. Rybakowski F, Slopien A, Dmitrzak-Weglarz M, Czerski P, Rajewski A, et al. (2006) The 5-HT2A −1438 A/G and 5-HTTLPR polymorphisms and personality dimensions in adolescent anorexia nervosa: association study. Neuropsychobiology 53: 33–39. [DOI] [PubMed] [Google Scholar]
- 42.Farber L, Haus U, Spath M, Drechsler S (2004) Physiology and pathophysiology of the 5-HT3 receptor. Scand J Rheumatol Suppl 119: 2–8. [PubMed]
- 43. Need AC, Ahmadi KR, Spector TD, Goldstein DB (2006) Obesity is associated with genetic variants that alter dopamine availability. Ann Hum Genet 70: 293–303. [DOI] [PubMed] [Google Scholar]
- 44. Fuemmeler BF, Agurs-Collins TD, McClernon FJ, Kollins SH, Kail ME, et al. (2008) Genes implicated in serotonergic and dopaminergic functioning predict BMI categories. Obesity (Silver Spring) 16: 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fisher RA (1918) The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh 52: 399–433. [Google Scholar]
- 46. Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, et al. (2012) Genetic contributions to stability and change in intelligence from childhood to old age. Nature 482: 212–215. [DOI] [PubMed] [Google Scholar]
- 48. Davies G, Tenesa A, Payton A, Yang J, Harris SE, et al. (2011) Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 16: 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen C, Moyzis R, Stern H, He Q, Li H, et al. (2011) Contributions of dopamine-related genes and environmental factors to highly sensitive personality: a multi-step neuronal system-level approach. PLoS One 6: e21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen C, Moyzis R, He Q, Li H, Li J, et al. (2012) Genetic variations in the dopaminergic system and alcohol use: a system-level analysis. Addict Biol 17: 479–489. [DOI] [PubMed] [Google Scholar]
- 51. Zhu B, Chen C, Moyzis RK, Dong Q, He Q, et al. (2012) Genetic variations in the dopamine system and facial expression recognition in healthy chinese college students. Neuropsychobiology 65: 83–89. [DOI] [PubMed] [Google Scholar]
- 52. Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M (1993) Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption–II. Addiction 88: 791–804. [DOI] [PubMed] [Google Scholar]
- 53. Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO (1991) The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict 86: 1119–1127. [DOI] [PubMed] [Google Scholar]
- 54. Lim JS, Son HK, Park SK, Jacobs DR Jr, Lee DH (2011) Inverse associations between long-term weight change and serum concentrations of persistent organic pollutants. Int J Obes (Lond) 35: 744–747. [DOI] [PubMed] [Google Scholar]
- 55.Kuczmarski MF, Kuczmarski RJ, Najjar M (2001) Effects of age on validity of self-reported height, weight, and body mass index: findings from the Third National Health and Nutrition Examination Survey, 1988–1994. J Am Diet Assoc 101: 28–34; quiz 35–26. [DOI] [PubMed]
- 56. Goodman E, Hinden BR, Khandelwal S (2000) Accuracy of teen and parental reports of obesity and body mass index. Pediatrics 106: 52–58. [DOI] [PubMed] [Google Scholar]
- 57. Strauss RS (1999) Comparison of measured and self-reported weight and height in a cross-sectional sample of young adolescents. Int J Obes Relat Metab Disord 23: 904–908. [DOI] [PubMed] [Google Scholar]
- 58. The International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- 59. Sabol SZ, Hu S, Hamer D (1998) A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet 103: 273–279. [DOI] [PubMed] [Google Scholar]
- 60. Qian Q, Wang Y, Zhou R, Li J, Wang B, et al. (2003) Family-based and case-control association studies of catechol-O-methyltransferase in attention deficit hyperactivity disorder suggest genetic sexual dimorphism. Am J Med Genet B Neuropsychiatr Genet 118B: 103–109. [DOI] [PubMed] [Google Scholar]
- 61. Collier DA, Stober G, Li T, Heils A, Catalano M, et al. (1996) A novel functional polymorphism within the promoter of the serotonin transporter gene: possible role in susceptibility to affective disorders. Mol Psychiatry 1: 453–460. [PubMed] [Google Scholar]
- 62. Guan L, Wang B, Chen Y, Yang L, Li J, et al. (2009) A high-density single-nucleotide polymorphism screen of 23 candidate genes in attention deficit hyperactivity disorder: suggesting multiple susceptibility genes among Chinese Han population. Mol Psychiatry 14: 546–554. [DOI] [PubMed] [Google Scholar]
- 63. Wang ET, Kodama G, Baldi P, Moyzis RK (2006) Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci U S A 103: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK (2007) Recent acceleration of human adaptive evolution. Proc Natl Acad Sci U S A 104: 20753–20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, et al. (2010) Data quality control in genetic case-control association studies. Nat Protoc 5: 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sakamaki R, Toyama K, Amamoto R, Liu C-J, Shinfuku N (2005) Nutritional knowledge, food habits and health attitude of Chinese university students -a cross sectional study. Nutrition Journal 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ge K (1997) Body mass index of young Chinese adults. Asia Pacific J Clin Nutr 6: 175–179. [PubMed] [Google Scholar]
- 69. Lei SF, Liu MY, Chen XD, Deng FY, Lv JH, et al. (2006) Relationship of total body fatness and five anthropometric indices in Chinese aged 20–40 years: different effects of age and gender. Eur J Clin Nutr 60: 511–518. [DOI] [PubMed] [Google Scholar]
- 70. Erickson JD, Schafer MK, Bonner TI, Eiden LE, Weihe E (1996) Distinct pharmacological properties and distribution in neurons and endocrine cells of two isoforms of the human vesicular monoamine transporter. Proc Natl Acad Sci U S A 93: 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mahata SK, Mahata M, Fischer-Colbrie R, Winkler H (1993) Vesicle monoamine transporters 1 and 2: differential distribution and regulation of their mRNAs in chromaffin and ganglion cells of rat adrenal medulla. Neurosci Lett 156: 70–72. [DOI] [PubMed] [Google Scholar]
- 72. Peter D, Liu Y, Sternini C, de Giorgio R, Brecha N, et al. (1995) Differential expression of two vesicular monoamine transporters. J Neurosci 15: 6179–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hayashi M, Haga M, Yatsushiro S, Yamamoto A, Moriyama Y (1999) Vesicular monoamine transporter 1 is responsible for storage of 5-hydroxytryptamine in rat pinealocytes. J Neurochem 73: 2538–2545. [DOI] [PubMed] [Google Scholar]
- 74. Agil A, Navarro-Alarcon M, Ruiz R, Abuhamadah S, El-Mir MY, et al. (2011) Beneficial effects of melatonin on obesity and lipid profile in young Zucker diabetic fatty rats. J Pineal Res 50: 207–212. [DOI] [PubMed] [Google Scholar]
- 75. Wolden-Hanson T, Mitton DR, McCants RL, Yellon SM, Wilkinson CW, et al. (2000) Daily melatonin administration to middle-aged male rats suppresses body weight, intraabdominal adiposity, and plasma leptin and insulin independent of food intake and total body fat. Endocrinology 141: 487–497. [DOI] [PubMed] [Google Scholar]
- 76. Tan DX, Manchester LC, Fuentes-Broto L, Paredes SD, Reiter RJ (2011) Significance and application of melatonin in the regulation of brown adipose tissue metabolism: relation to human obesity. Obes Rev 12: 167–188. [DOI] [PubMed] [Google Scholar]
- 77. Lohoff FW, Lautenschlager M, Mohr J, Ferraro TN, Sander T, et al. (2008) Association between variation in the vesicular monoamine transporter 1 gene on chromosome 8p and anxiety-related personality traits. Neurosci Lett 434: 41–45. [DOI] [PubMed] [Google Scholar]
- 78. Jorm AF, Korten AE, Christensen H, Jacomb PA, Rodgers B, et al. (2003) Association of obesity with anxiety, depression and emotional well-being: a community survey. Aust N Z J Public Health 27: 434–440. [DOI] [PubMed] [Google Scholar]
- 79. Strine TW, Mokdad AH, Dube SR, Balluz LS, Gonzalez O, et al. (2008) The association of depression and anxiety with obesity and unhealthy behaviors among community-dwelling US adults. Gen Hosp Psychiatry 30: 127–137. [DOI] [PubMed] [Google Scholar]
- 80. Zhao G, Ford ES, Dhingra S, Li C, Strine TW, et al. (2009) Depression and anxiety among US adults: associations with body mass index. Int J Obes (Lond) 33: 257–266. [DOI] [PubMed] [Google Scholar]
- 81. Furnes MW, Zhao CM, Stenstrom B, Arum CJ, Tommeras K, et al. (2009) Feeding behavior and body weight development: lessons from rats subjected to gastric bypass surgery or high-fat diet. J Physiol Pharmacol 60 Suppl 7: 25–31. [PubMed] [Google Scholar]
- 82. Zhao CM, Furnes MW, Stenstrom B, Kulseng B, Chen D (2008) Characterization of obestatin- and ghrelin-producing cells in the gastrointestinal tract and pancreas of rats: an immunohistochemical and electron-microscopic study. Cell Tissue Res 331: 575–587. [DOI] [PubMed] [Google Scholar]
- 83. Thompson AJ, Lummis SC (2007) The 5-HT3 receptor as a therapeutic target. Expert Opin Ther Targets 11: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen CY, Asakawa A, Fujimiya M, Lee SD, Inui A (2009) Ghrelin gene products and the regulation of food intake and gut motility. Pharmacol Rev 61: 430–481. [DOI] [PubMed] [Google Scholar]
- 85. Yamada K, Hattori E, Iwayama Y, Ohnishi T, Ohba H, et al. (2006) Distinguishable haplotype blocks in the HTR3A and HTR3B region in the Japanese reveal evidence of association of HTR3B with female major depression. Biol Psychiatry 60: 192–201. [DOI] [PubMed] [Google Scholar]
- 86. Kato M, Fukuda T, Wakeno M, Fukuda K, Okugawa G, et al. (2006) Effects of the serotonin type 2A, 3A and 3B receptor and the serotonin transporter genes on paroxetine and fluvoxamine efficacy and adverse drug reactions in depressed Japanese patients. Neuropsychobiology 53: 186–195. [DOI] [PubMed] [Google Scholar]
- 87. Sugai T, Suzuki Y, Sawamura K, Fukui N, Inoue Y, et al. (2006) The effect of 5-hydroxytryptamine 3A and 3B receptor genes on nausea induced by paroxetine. Pharmacogenomics J 6: 351–356. [DOI] [PubMed] [Google Scholar]
- 88. De Luca V, Mueller DJ, de Bartolomeis A, Kennedy JL (2007) Association of the HTR2C gene and antipsychotic induced weight gain: a meta-analysis. Int J Neuropsychopharmacol 10: 697–704. [DOI] [PubMed] [Google Scholar]
- 89. Reynolds GP, Hill MJ, Kirk SL (2006) The 5-HT2C receptor and antipsychoticinduced weight gain - mechanisms and genetics. J Psychopharmacol 20: 15–18. [DOI] [PubMed] [Google Scholar]
- 90. Cowen PJ, Clifford EM, Walsh AE, Williams C, Fairburn CG (1996) Moderate dieting causes 5-HT2C receptor supersensitivity. Psychol Med 26: 1155–1159. [DOI] [PubMed] [Google Scholar]
- 91. Collier DA, Arranz MJ, Li T, Mupita D, Brown N, et al. (1997) Association between 5-HT2A gene promoter polymorphism and anorexia nervosa. Lancet 350: 412. [DOI] [PubMed] [Google Scholar]
- 92. Enoch MA, Kaye WH, Rotondo A, Greenberg BD, Murphy DL, et al. (1998) 5-HT2A promoter polymorphism −1438G/A, anorexia nervosa, and obsessive-compulsive disorder. Lancet 351: 1785–1786. [DOI] [PubMed] [Google Scholar]
- 93. Nakamura T, Matsushita S, Nishiguchi N, Kimura M, Yoshino A, et al. (1999) Association of a polymorphism of the 5HT2A receptor gene promoter region with alcohol dependence. Mol Psychiatry 4: 85–88. [DOI] [PubMed] [Google Scholar]
- 94. Ricca V, Nacmias B, Boldrini M, Cellini E, di Bernardo M, et al. (2004) Psychopathological traits and 5-HT2A receptor promoter polymorphism (−1438 G/A) in patients suffering from Anorexia Nervosa and Bulimia Nervosa. Neurosci Lett 365: 92–96. [DOI] [PubMed] [Google Scholar]
- 95. Halder I, Muldoon MF, Ferrell RE, Manuck SB (2007) Serotonin Receptor 2A (HTR2A) Gene Polymorphisms Are Associated with Blood Pressure, Central Adiposity, and the Metabolic Syndrome. Metab Syndr Relat Disord 5: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rosmond R, Bouchard C, Bjorntorp P (2002) 5-HT2A receptor gene promoter polymorphism in relation to abdominal obesity and cortisol. Obes Res 10: 585–589. [DOI] [PubMed] [Google Scholar]
- 97. Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lemmens VE, Oenema A, Klepp KI, Henriksen HB, Brug J (2008) A systematic review of the evidence regarding efficacy of obesity prevention interventions among adults. Obes Rev 9: 446–455. [DOI] [PubMed] [Google Scholar]
- 99. Anderson JW, Konz EC, Frederich RC, Wood CL (2001) Long-term weight-loss maintenance: a meta-analysis of US studies. Am J Clin Nutr 74: 579–584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed information of the loci used in this study.
(DOC)
Means and standard deviations of BMI for each polymorphism, and main effects and post hoc comparisons of each locus.
(DOC)