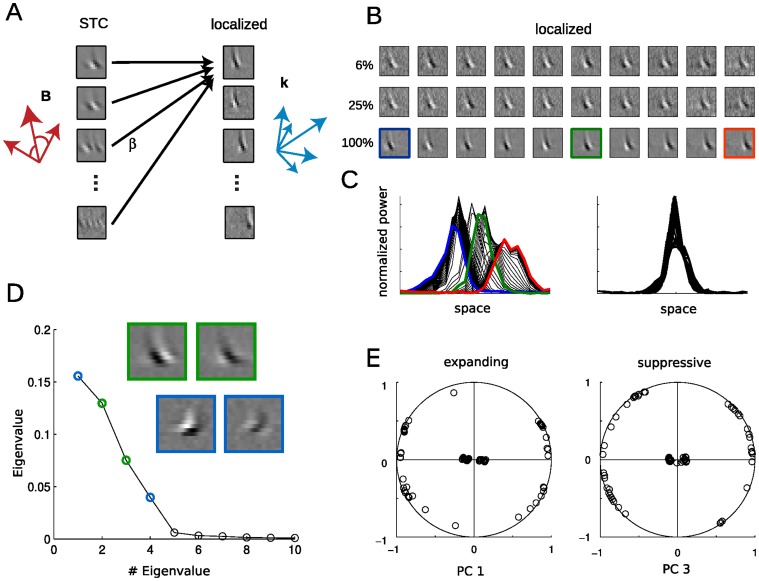
Figure 2. Localized filters provide a biologically plausible alternative description of the STC subspace.
A. The subspace defined by the STC filters can be used to find a set of localized filters, where each localized filter (right) is a linear combination of the set of STC filters (left). The small filters (STC and localized) are taken from 1B and 2B (last row) and exemplify the different features that can be used to represent a cells sensitivity. Red arrows illustrate an orthogonal vector space defined by the STC filters, while blue arrows illustrate a larger number of non-orthogonal localized filters that span the same subspace. B. Localization detects a large number of filters within the STC subspace, and 10 (from 47) localized solutions spanning the RF are shown. As more data is used for the analysis, the properties of the filters remain relatively constant, but their features become less polluted by noise. The localized solutions show similar spatiotemporal sensitivity, with the main differences between the filters amounting to differences in position and phase. C. The power profiles of 47 localized excitatory filters (left) have been aligned (right) to compare the similarity of their spatial envelopes. D. The 47 excitatory and 47 suppressive localized solutions form 2 homogeneous populations of spatially confined features that are succinctly described by four eigenvectors - two spanning the excitatory subspace, and two spanning the suppressive subspace - as determined by PCA applied to the set of spatially shifted localized filters in D. E. The filters are well represented by these PCA eigenvectors (98% exp, 97% suppressive) and evenly cover the range of possible combinations of these vectors.