Abstract
Alu repeats are short interspersed elements (SINEs) of dimeric structure whose transposition sometimes leads to heritable disorders in humans. Human cells contain a poly(A)- small cytoplasmic transcript of -120 nucleotides (nt) homologous to the left Alu monomer. Although its monomeric size indicates that small cytoplasmic Alu (scAlu) RNA is not an intermediary of human Alu transpositions, a less abundant poly(A)-containing Alu transcript of dimeric size and specificity expected of a transposition intermediary is also detectable in HeLa cells (A. G. Matera, U. Hellmann, M. F. Hintz, and C. W. Schmid, Mol. Cell. Biol. 10:5424-5432, 1990). Although its function is unknown, the accumulation of Alu RNA and its ability to interact with a conserved protein suggest a role in cell biology (D.-Y. Chang and R. J. Maraia, J. Biol. Chem. 268:6423-28, 1993). The relationship between the -120- and -300-nt Alu transcripts had not been determined. However, a B1 SINE produces scB1 RNA by posttranscriptional processing, suggesting a similar pathway for scAlu. An Alu SINE which recently transposed into the neurofibromatosis 1 locus was expressed in microinjected frog oocytes. This neurofibromatosis 1 Alu produced a primary transcript followed by the appearance of the scAlu species. 3' processing of a synthetic -300-nt Alu RNA by HeLa nuclear extract in vitro also produced scAlu RNA. Primer extension of scAlu RNA indicates synthesis by RNA polymerase III. HeLa-derived scAlu cDNAs were cloned so as to preserve their 5'-terminal sequences and were found to correspond to polymerase III transcripts of the left monomeric components of three previously identified Alu SINE subfamilies. Rodent x human somatic cell hybrids express Alu RNAs whose size, heterogeneous length, and chromosomal distribution indicate their derivation from SINEs. The coexpression of dimeric and monomeric Alu RNA in several hybrids suggests a precursor-product relationship.
Full text
PDF



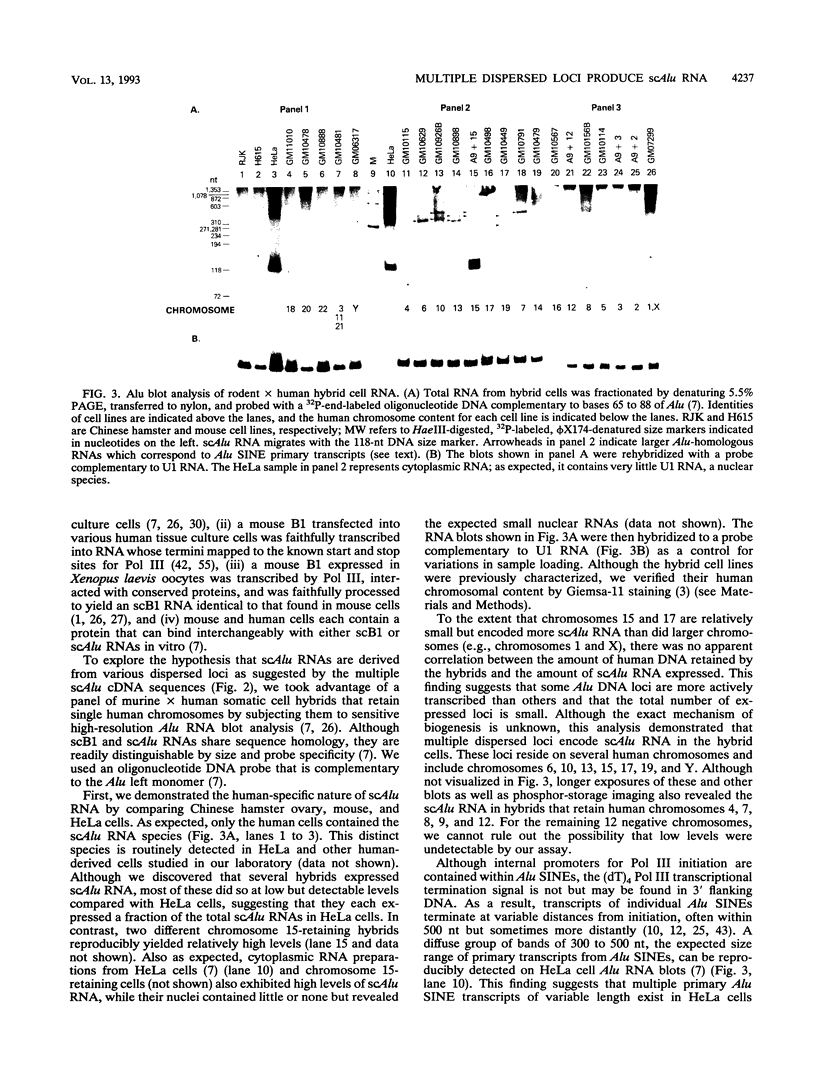
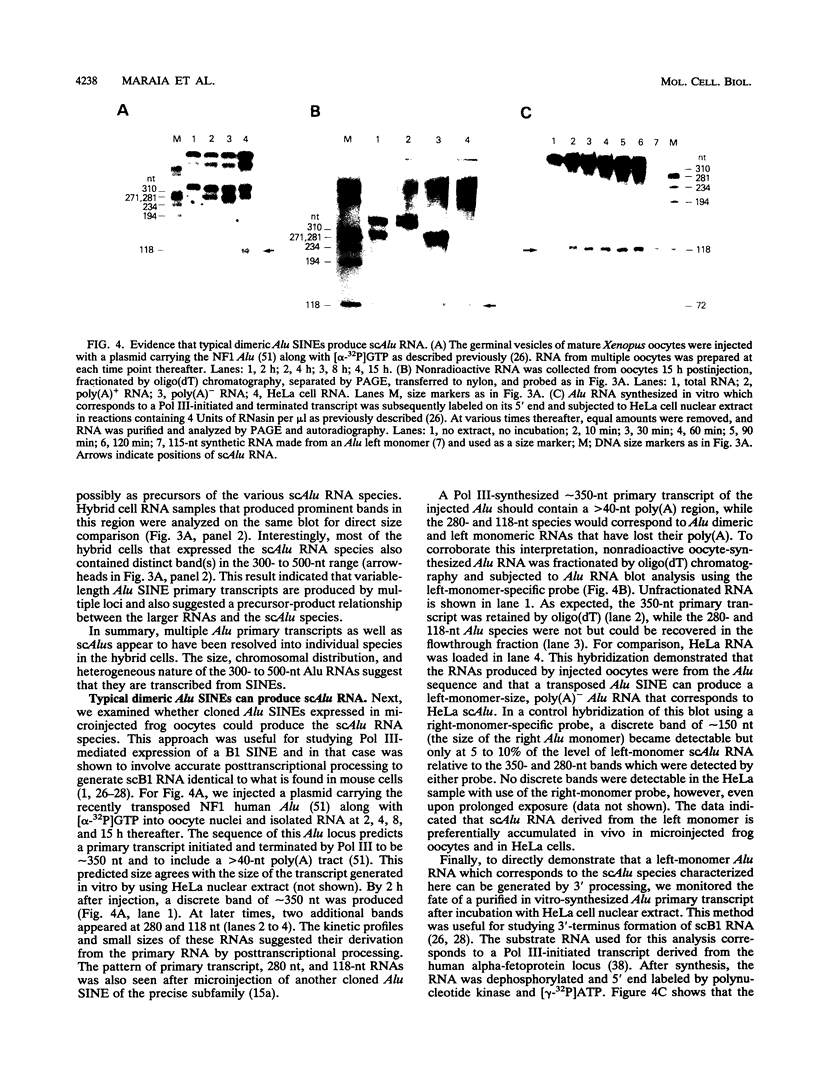



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adeniyi-Jones S., Zasloff M. Transcription, processing and nuclear transport of a B1 Alu RNA species complementary to an intron of the murine alpha-fetoprotein gene. Nature. 1985 Sep 5;317(6032):81–84. doi: 10.1038/317081a0. [DOI] [PubMed] [Google Scholar]
- Batzer M. A., Kilroy G. E., Richard P. E., Shaikh T. H., Desselle T. D., Hoppens C. L., Deininger P. L. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 1990 Dec 11;18(23):6793–6798. doi: 10.1093/nar/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrow M., Cross J. Differential staining of human and mouse chromosomes in interspecific cell hybrids. Nature. 1974 Sep 6;251(5470):77–79. doi: 10.1038/251077a0. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Gould S. J. On "genomenclature": a comprehensive (and respectful) taxonomy for pseudogenes and other "junk DNA". Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Retroposons--seeds of evolution. Science. 1991 Feb 15;251(4995):753–753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Chang D. Y., Maraia R. J. A cellular protein binds B1 and Alu small cytoplasmic RNAs in vitro. J Biol Chem. 1993 Mar 25;268(9):6423–6428. [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Characterization of a third major SINE family of repetitive sequences in the galago genome. Nucleic Acids Res. 1991 Apr 11;19(7):1649–1656. doi: 10.1093/nar/19.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou-Pachnis A., Tsichlis P. N. Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res. 1985 Dec 9;13(23):8379–8387. doi: 10.1093/nar/13.23.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. T., Pan J., Duncan C. H., Weissman S. M. Transcriptional analysis of interspersed repetitive polymerase III transcription units in human DNA. Nucleic Acids Res. 1981 Mar 11;9(5):1171–1189. doi: 10.1093/nar/9.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman S. A., Deininger P. L., LaPorte P., Friedmann T., Geiduschek E. P. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981 Dec 11;9(23):6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Collins K. W., Hernandez E. M., Garrard W. T. Vacuum blotting: a simple method for transferring DNA from sequencing gels to nylon membranes. Gene. 1988 Dec 30;74(2):347–356. doi: 10.1016/0378-1119(88)90168-0. [DOI] [PubMed] [Google Scholar]
- Hess J., Perez-Stable C., Wu G. J., Weir B., Tinoco I., Jr, Shen C. K. End-to-end transcription of an Alu family repeat. A new type of polymerase-III-dependent terminator and its evolutionary implication. J Mol Biol. 1985 Jul 5;184(1):7–21. doi: 10.1016/0022-2836(85)90039-7. [DOI] [PubMed] [Google Scholar]
- Howard B. H., Sakamoto K. Alu interspersed repeats: selfish DNA or a functional gene family? New Biol. 1990 Sep;2(9):759–770. [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Jurka J., Milosavljevic A. Reconstruction and analysis of human Alu genes. J Mol Evol. 1991 Feb;32(2):105–121. doi: 10.1007/BF02515383. [DOI] [PubMed] [Google Scholar]
- Jurka J., Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Zuckerkandl E. Free left arms as precursor molecules in the evolution of Alu sequences. J Mol Evol. 1991 Jul;33(1):49–56. doi: 10.1007/BF02100195. [DOI] [PubMed] [Google Scholar]
- Labuda D., Sinnett D., Richer C., Deragon J. M., Striker G. Evolution of mouse B1 repeats: 7SL RNA folding pattern conserved. J Mol Evol. 1991 May;32(5):405–414. doi: 10.1007/BF02101280. [DOI] [PubMed] [Google Scholar]
- Labuda D., Striker G. Sequence conservation in Alu evolution. Nucleic Acids Res. 1989 Apr 11;17(7):2477–2491. doi: 10.1093/nar/17.7.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang E. P., Liu W. M., Hashimoto C., Choudary P. V., Schmid C. W. Phylogenetic evidence for multiple Alu source genes. J Mol Evol. 1992 Jul;35(1):7–16. doi: 10.1007/BF00160256. [DOI] [PubMed] [Google Scholar]
- Lin C. S., Goldthwait D. A., Samols D. Identification of Alu transposition in human lung carcinoma cells. Cell. 1988 Jul 15;54(2):153–159. doi: 10.1016/0092-8674(88)90547-8. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Colozzo M. T. Synthesis in vitro of an exceptionally long RNA transcript promoted by an AluI sequence. Nature. 1982 Nov 25;300(5890):376–379. doi: 10.1038/300376a0. [DOI] [PubMed] [Google Scholar]
- Maraia R. J., Chang D. Y., Wolffe A. P., Vorce R. L., Hsu K. The RNA polymerase III terminator used by a B1-Alu element can modulate 3' processing of the intermediate RNA product. Mol Cell Biol. 1992 Apr;12(4):1500–1506. doi: 10.1128/mcb.12.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R. J. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res. 1991 Oct 25;19(20):5695–5702. doi: 10.1093/nar/19.20.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R., Zasloff M., Plotz P., Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol Cell Biol. 1988 Oct;8(10):4433–4440. doi: 10.1128/mcb.8.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Hellmann U., Hintz M. F., Schmid C. W. Recently transposed Alu repeats result from multiple source genes. Nucleic Acids Res. 1990 Oct 25;18(20):6019–6023. doi: 10.1093/nar/18.20.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Hellmann U., Schmid C. W. A transpositionally and transcriptionally competent Alu subfamily. Mol Cell Biol. 1990 Oct;10(10):5424–5432. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G. A., Labuda D., Fontaine G., Saudubray J. M., Bonnefont J. P., Lyonnet S., Brody L. C., Steel G., Obie C., Valle D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. H., Baralle F. E. Directed semisynthetic point mutational analysis of an RNA polymerase III promoter. Nucleic Acids Res. 1983 Nov 25;11(22):7695–7700. doi: 10.1093/nar/11.22.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y., Lovell M., Taylor L., Pereira-Smith O. M. Isolation of monochromosomal hybrids following fusion of human diploid fibroblast-derived microcells with mouse A9 cells. Cytogenet Cell Genet. 1992;60(1):79–80. doi: 10.1159/000133300. [DOI] [PubMed] [Google Scholar]
- Quentin Y. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res. 1992 Feb 11;20(3):487–493. doi: 10.1093/nar/20.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res. 1992 Jul 11;20(13):3397–3401. doi: 10.1093/nar/20.13.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. Successive waves of fixation of B1 variants in rodent lineage history. J Mol Evol. 1989 Apr;28(4):299–305. doi: 10.1007/BF02103425. [DOI] [PubMed] [Google Scholar]
- Quentin Y. The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol. 1988;27(3):194–202. doi: 10.1007/BF02100074. [DOI] [PubMed] [Google Scholar]
- Ryan S. C., Dugaiczyk A. Newly arisen DNA repeats in primate phylogeny. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9360–9364. doi: 10.1073/pnas.86.23.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. W. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 1991 Oct 25;19(20):5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C., Maraia R. Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Genet Dev. 1992 Dec;2(6):874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- Scott R. W., Tilghman S. M. Transient expression of a mouse alpha-fetoprotein minigene: deletion analyses of promoter function. Mol Cell Biol. 1983 Jul;3(7):1295–1309. doi: 10.1128/mcb.3.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. The organization, structure, and in vitro transcription of Alu family RNA polymerase III transcription units in the human alpha-like globin gene cluster: precipitation of in vitro transcripts by lupus anti-La antibodies. J Mol Appl Genet. 1982;1(4):343–360. [PubMed] [Google Scholar]
- Shen M. R., Batzer M. A., Deininger P. L. Evolution of the master Alu gene(s). J Mol Evol. 1991 Oct;33(4):311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Sinnett D., Richer C., Deragon J. M., Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J Biol Chem. 1991 May 15;266(14):8675–8678. [PubMed] [Google Scholar]
- Sinnett D., Richer C., Deragon J. M., Labuda D. Alu RNA transcripts in human embryonal carcinoma cells. Model of post-transcriptional selection of master sequences. J Mol Biol. 1992 Aug 5;226(3):689–706. doi: 10.1016/0022-2836(92)90626-u. [DOI] [PubMed] [Google Scholar]
- Slagel V., Flemington E., Traina-Dorge V., Bradshaw H., Deininger P. Clustering and subfamily relationships of the Alu family in the human genome. Mol Biol Evol. 1987 Jan;4(1):19–29. doi: 10.1093/oxfordjournals.molbev.a040422. [DOI] [PubMed] [Google Scholar]
- Strub K., Moss J., Walter P. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol Cell Biol. 1991 Aug;11(8):3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. R., Andersen L. B., Saulino A. M., Gregory P. E., Glover T. W., Collins F. S. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991 Oct 31;353(6347):864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- Watson J. B., Sutcliffe J. G. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol Cell Biol. 1987 Sep;7(9):3324–3327. doi: 10.1128/mcb.7.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Willard C., Nguyen H. T., Schmid C. W. Existence of at least three distinct Alu subfamilies. J Mol Evol. 1987;26(3):180–186. doi: 10.1007/BF02099850. [DOI] [PubMed] [Google Scholar]
- Young P. R., Scott R. W., Hamer D. H., Tilghman S. M. Construction and expression in vivo of an internally deleted mouse alpha-fetoprotein gene: presence of a transcribed Alu-like repeat within the first intervening sequence. Nucleic Acids Res. 1982 May 25;10(10):3099–3116. doi: 10.1093/nar/10.10.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Latter G., Jurka J. Maintenance of function without selection: Alu sequences as "cheap genes". J Mol Evol. 1989 Dec;29(6):504–512. doi: 10.1007/BF02602922. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]