Figure 1. Engineering monobodies to target the Erk-2 CD domain.
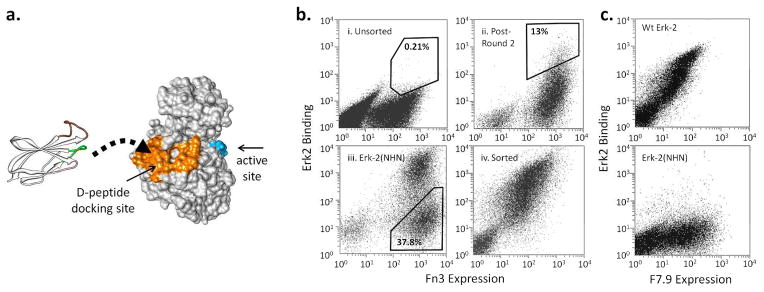
a. The Erk-2 residues within 5 Å of bound D-peptide (2GPH) are colored orange. The activation loop residues are colored in blue to show the separation between the docking domain and the active site. An Fn3 structure is shown to the scale on the left and is depicted as binding the docking surface. The randomized loops are shown in color: BC (green), DE (pink), FG (brown).
b. The yeast displayed Fn3 library was sorted using streptavidin-coated magnetic beads and FACS. During FACS, the cells were labeled with a cMyc antibody and Erk-2 to normalize binding with protein expression. i) Unsorted cells labeled with 1 μM of Erk-2. ii) Cells that have been sorted twice by FACS and then labeled with 250 nM of Erk-2. iii) Cells labeled with Erk-2(NHN) during negative sorting (Round 6). iv) Cells after the final round of FACS labeled with 10 nM of Erk-2. The percentages indicate the fraction of the cells within the indicated gates.
c. A representative selected clone, F7.9, binds 10 nM wt Erk-2 but not Erk-2(NHN). The lack of binding to the mutant indicates that the monobody epitope includes one or more of the mutated residues.
