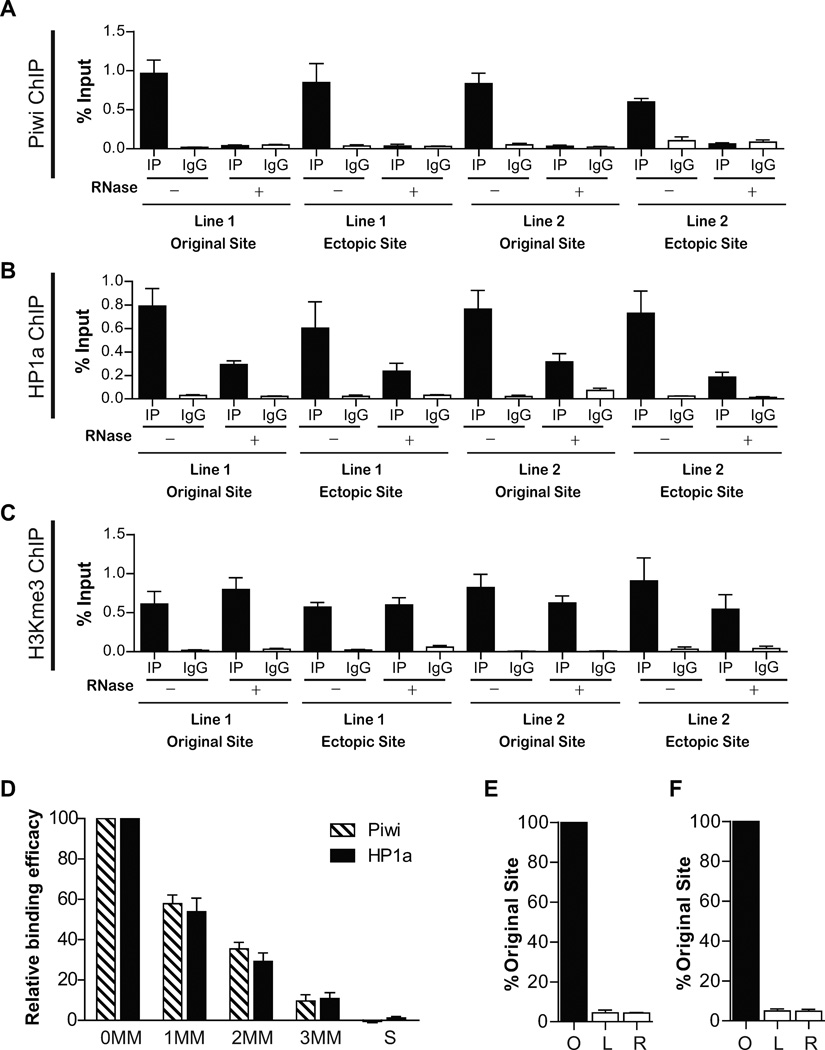
Figure 4. The ectopic recruitment of Piwi and HP1a is sensitive to RNase treatment and is achieved by piRNA from the original site.
(A) Piwi ChIP in the presence or absence of RNase A digestion. In both Line 1 and Line 2, Piwi binding to the original site and the ectopic site is abolished by RNase treatment. Error bars in A-F denote S.D.
(B) HP1a ChIP in the presence or absence of RNase A. Similar to (A), HP1a binding to original site and the ectopic site is impaired in both Line1 and Line 2 by RNase treatment.
(C) H3K9me3 ChIP in the presence or absence of RNase A. Unlike (A) and (B), H3K9me3 signal in the original site and the ectopic site is not affected in both Line1 and Line 2 by RNase treatment.
(D) The impact of mismatches (MM) on the efficacy of piRNA biding to its target sequence. S: scrambled sequence. See also Figure S3.
(E–F) The ectopic sites in Line 1 (C) and Line 2 (D) produce negligible levels of RNA transcripts comparing to their original piRNA-generating sites. O: original site; L and R: sequences spanning the left and right insertion junctions at the ectopic site.