Abstract
After the worldwide cholera epidemic in 1993, permanent environmental monitoring of hydrographic basins was established in Pernambuco, Brazil, where cholera is endemic. After a quiescent period, 4 rfbN (serogroup O1) positive water samples that were culture negative were detected by multiplex single-tube nested PCR (MSTNPCR); 2 of these were also ctxA (cholera toxin) positive. From May to June 2012, 30 V. cholerae O1 isolates were obtained by culturing samples. These isolates were analyzed for the presence of virulence genes by PCR, intergenic spacer region 16S-23S PCR (ISR-PCR), and pulsed field gel electrophoresis (PFGE). The isolates were positive for the rfbN gene and negative for the assessed pathogenic genes and were classified into 2 groups by ISR and the same profile by PFGE. Close genetic similarity was observed between them (2012) and environmental strains from 2004 to 2005, indicating the permanence of endemic V. cholerae O1 in the region.
1. Introduction
Vibrio cholerae has played a prominent role in human history and has caused several epidemics that caused many deaths worldwide. While more than 200 O serogroups of V. cholerae have been identified, only the O1 and O139 serogroups have been associated with epidemics [1]. Other serogroups commonly known as non-O1/non-O139 [2] coexist in the environment with O1 strains [3]. In Brazil, the O1 serogroup has been recognized as the causative agent of past epidemics, but it is likely that other serogroups were involved in recent small outbreaks [4, 5].
Since the last world cholera epidemic in 1993 [6], the Pernambuco (PE) state Department of Health (Secretaria Estadual de Saúde (SES/PE)) has established permanent environmental monitoring of the hydrographic basins in the state for the detection of V. cholerae [7]. This investigation was first based on the standard culture procedures of water samples, and later a molecular technique, multiplex single-tube nested PCR (MSTNPCR), is used to target the ctxA gene, which encodes cholera toxin subunit A, and the rfbN gene, which is specific to serogroup O1 [8]. In May 2012, four rfbN positive but not culturable water samples, of which two were ctxA (cholera toxin) positive, were detected by MSTNPCR. After that, the monitoring was intensified and, from May to June 2012, 30 V. cholerae O1-positive cultures were unexpectedly isolated from four hydrographic basins.
2. Methods
2.1. Environmental Water Sample Collection and Bacteriological Analysis
The Moore swab technique [9] was used to collect environmental water samples from hydrographic basins, and the samples were submitted to enrichment in alkaline peptone water (APW). The samples were streaked on thiosulfate-citrate-bile-sucrose agar (TCBS) plates and incubated overnight at 37°C. Yellow colonies were tested for the presence of oxidase with oxidase strips and inoculated in Instituto Adolfo Lutz (IAL)/Rugai tube [10] for motility, glucose and lactose fermentation, gas and H2S production, phenylalanine, urea, indol, and lysine tests. The O serogroups were determined by a slide agglutination test with polyvalent O1 and O139 and monospecific Inaba, and Ogawa antisera [11].
2.2. Molecular Analysis
The isolates were analyzed for the presence of V. cholerae virulence genes (ctxA, ctxB, tcpA, ace, and zot), the serogroup O1-specific gene (rfbN), by intergenic spacer region 16S–23S (ISR-PCR) PCR amplification, and pulsed-field gel electrophoresis (PFGE). Vibrio cholerae O1 ATCC 569B was used as control.
2.3. DNA Extraction and PCR Reactions
The genomic DNA was extracted by a heat soak DNA extraction procedure based on Keim et al. [12]. One colony grown on BHI plates was suspended in 200 μL of TE 10 : 1 (10 mM Tris-HCl pH 8.0, 1 mM EDTA), boiled for 20 minutes, and immediately used in PCR reactions with primers designed for the amplification of ctxA and tcpA [13], zot and ace [14], ctxB [15], and rfbN [16]. Each PCR reaction consisted of 50 mM KCl, 10 mM Tris-HCl, 2.5 mM MgCl2, 400 mM each dNTP, 20 pmol primers, 1 U of Taq DNA polymerase (Promega), heat soak DNA sample (10 μL), and sterile water in a final volume of 25 μL. ISR-PCR was performed as previously described [17].
The PCR reactions were conducted in a Biometra T-3000 Genetic Analyzer thermal cycler using standard procedures. The products were electrophoresed in 1% agarose gels containing SYBR Safe DNA gel stain (Invitrogen) and photographed with Kodak 1D Image Analysis software, version 3.5 (Digital Kodak Science).
2.4. PFGE
The typing of V. cholerae isolates was performed by PFGE according to a PulseNet [18] standardized protocol. NotI-HF (New England Biolabs) digested DNA fragments were separated in a CHEF-DR III Bio-Rad system (Contour-Clamped Homogeneous Electric Fields/Bio-Rad, Hercules, CA, USA) in 1% SeaKem Gold agarose (Lonza, Rockland, ME, USA) gels in 0.5% TBE running buffer at 14°C, with a ramping time of 4.5 V/cm for 22 h. The comparisons in the study included one non-O1 isolate (Vc479-1), which was obtained by coproculture from a diarrheic patient in 2012, V. cholerae O1 569B ATCC, and three O1 isolates from previous cholera outbreaks in Brazil (Vc460/04, Vc461/04, and Vc499/05). The Lambda PFGE marker (New England Biolabs, Country RD Ipswich, MA, USA) was the molecular weight standard. The ethidium bromide (1 µg/mL) stained bands were visualized under UV light, and images were captured by 1D Image Analysis Software, version 3.5 (Kodak Digital Science, New Haven, CT, USA).
The PFGE profiles were analyzed by visual inspection with NTSYSpc software version 2.11X [19]. The similarity among the samples was determined by Dice's similarity coefficient [20], and a dendrogram was constructed with UPGMA, following a profile cluster analysis as described by Tenover et al. [21].
3. Results
3.1. PCR
MSTNPCR analysis of water samples from aquatic basins of PE, Brazil, from May to June 2012 resulted in the detection of four rfbN-positive samples, of which two were also ctxA positive. The cultures of these samples were negative. Later on, from May to June 2012, 30 V. cholerae isolates were obtained by culture from four hydrographic basins (Figure 1) and classified as O1 by a slide agglutination test. All samples were positive for the rfbN gene characterizing the O1 serogroup, and none harbored the pathogenic genes evaluated by PCR.
Figure 1.
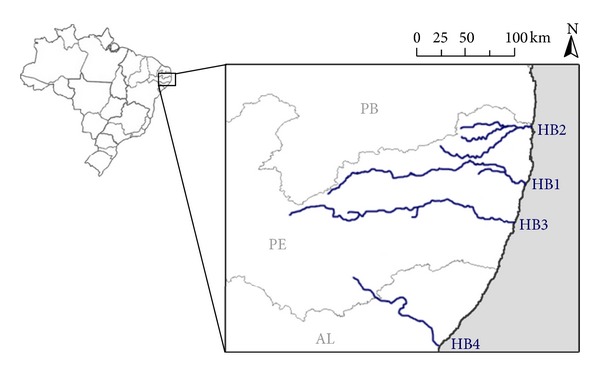
Map of the four hydrographic basins from which Vibrio cholerae O1 was isolated. HB1: Capibaribe hydrographic basin, HB2: Goiana hydrographic basin, HB3: Ipojuca hydrographic basin, HB4: Mundaú hydrographic basin, PE: Pernambuco state, PB: Paraíba state; AL: Alagoas state.
According to the ISR-PCR patterns generated, the 30 environmental V. cholerae O1 isolates were clustered into two distinct groups, named A and B. Group A included all isolates, with the exception of the two isolates (1880 and 1906) that belonged to group B.
3.2. PFGE
PFGE analysis of the 30 environmental V. cholerae O1 isolates, the non-O1 isolate from a diarrheic patient, the strains of V. cholerae O1 569B ATCC, and the previously isolated Brazilian strains Vc460/04, Vc461/04, and Vc499/05 grouped the 35 isolates into six clonally related pulsotypes (A–F).
The 30 V. cholerae O1 2012 environmental isolates were shown to be genetically identical, with a 0-1 band difference, and clustered into one pulsotype (A) that was subdivided in two (A1-A2) types. The three Brazilian V. cholerae O1 strains from 2004 (Vc460/04, Vc461/04) to 2005 (Vc499/05) were classified into three pulsotypes (B–D). The human isolate (non-O1, Vc479-1) formed a singleton (F), and the reference O1 strain Vc569B was the other singleton (E).
A close genetic relationship (~85% similarity) was observed among pulsotype A (2012 Brazilian environmental isolates) and pulsotypes B (a 2005 Brazilian environmental strain—Vc499/05) and C (a 2004 Brazilian environmental strain—Vc460/04), with differences in 4-5 bands (Figure 2).
Figure 2.
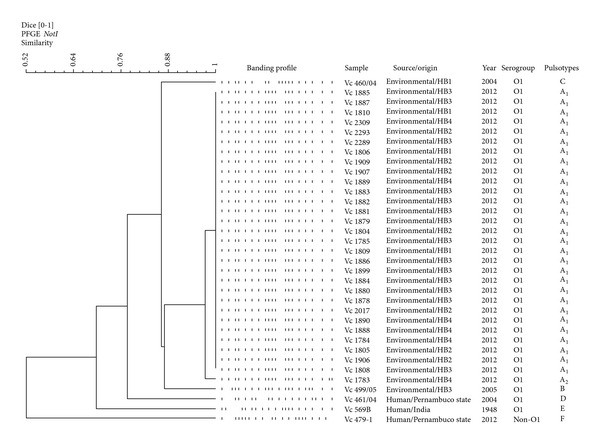
Dendrogram generated by Dice/UPGMA analysis (NTSYS v.2.11X, Applied Biostatistics) of PFGE NotI profiles of Vibrio cholerae isolates. HB1: Capibaribe hydrographic basin, HB2: Goiana hydrographic basin, HB3: Ipojuca hydrographic basin, HB4: Mundaú hydrographic basin.
4. Discussion
Through continuous surveillance for cholera in the hydrographic basins of PE, Brazil, four rfbN-positive samples, of which two were also ctxA positive, were uncovered by MSTNPCR. Although no cultures were isolated, this result suggests the presence of toxigenic V. cholerae strains and/or free phage in the aquatic environment of the region.
As recommended [22], this finding triggered prompt control measures by the SES/PE to halt further cholera outbreaks in the infected area. The measures included health education, drinking water treatment (the population was instructed to filter and treat drinking water with sodium hypochlorite, and to cook food thoroughly), and a compulsory search for V. cholerae in the environment and in cases of acute diarrhea. Thirty V. cholerae strains were further isolated by culture. They were classified as O1 by a slide agglutination test and were positive for the rfbN gene characterizing the O1 serogroup, but none harbored the pathogenic genes evaluated by PCR.
These results, together with the presence of nontoxigenic isolates in the same environment, are problematic because phage can infect V. cholerae O1 nontoxigenic isolates by the same way it infects other serogroups [5].
The close similarity among the isolates from the 2012, 2005, and 2004 events reveals the permanence of endemic V. cholerae O1 in the region. The slight divergences in their PFGE profiles are likely due to accumulated mutations resulting from the adaptive process of the bacteria to environmental and seasonal conditions. As anticipated, the Vc479-1 isolate from a diarrheic patient, although originated from the same place and time period as the cluster A samples, was genetically distinct (≥ differences 7) from the latter as it was revealed to be a non-O1 serogroup isolate. One O1 human 2004 isolate (Vc461/04) and the reference O1 strain (Vc569B) displayed unique pulsotypes and were isolated as singletons, most likely due to their specific toxigenic contents and different geographic origins.
In our study, isolates from different hydrographic basins displayed a clonal profile, possibly due to the presence of cyanobacteria, which are widespread in the region (PE State Department of Health, oral communication). According to Reidl and Klose [23], there is a direct relationship between cholera and algal blooms. It is likely that cyanobacteria are a contributing factor in the permanence of V. cholerae O1 in the aquatic environment, which depends on fecal contamination and/or environmental bacterial carriers [24].
In this paper, we report a sudden increase in V. cholerae O1 isolation in PE hydrographic basins from May to June 2012. The detection by MSTNPCR of four rfbN (serogroup O1)-positive culture-negative water samples, of which two were ctxA (cholera toxin) positive, suggests the presence of toxigenic V. cholerae strains and/or free phage in the environment. It is likely that phage infects toxigenic and nontoxigenic environmental strains by the same horizontal gene transfer mechanism by which it infects other serogroups. A genetic relationship was identified among V. cholerae O1 isolates. These findings indicate the possible emergence of pathogenic strains and the need for permanent monitoring of bacteria in the environment to avoid cholera outbreaks.
Conflict of Interests
The authors have no conflict of interests regarding this paper.
References
- 1.Goel AK, Jain M, Kumar P, Bhadauria S, Kmboj DV, Singh L. A new variant of Vibrio cholerae O1 El Tor causing cholera in India. Journal of Infection. 2008;57(3):280–281. [Google Scholar]
- 2.Safa A, Nair GB, Kong RYC. Evolution of new variants of Vibrio cholerae O1. Trends in Microbiology. 2010;18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee S, Ghosh K, Raychoudhuri A, et al. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. Journal of Clinical Microbiology. 2009;47(4):1087–1095. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theophilo GND, Rodrigues DP, Leal NC, Hofer E. Distribution of virulence markers in clinical and environmental Vibrio cholerae non-O1/non-O139 strains isolated in Brazil from 1991 to 2000. The Revista Do Instituto De Medicina Tropical De Sao Paulo. 2006;48:65–67. doi: 10.1590/s0036-46652006000200002. [DOI] [PubMed] [Google Scholar]
- 5.Cariri FAMO, Costa APR, Melo CC, et al. Characterization of potentially virulent non-O1/non-O139 Vibrio cholerae strains isolated from human patients. Clinical Microbiology and Infection. 2010;16(1):62–67. doi: 10.1111/j.1469-0691.2009.02763.x. [DOI] [PubMed] [Google Scholar]
- 6.Colwell RR. Global climate change and infectious diseases: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 7.Leal NC, de Araújo Figueiroa AC, Cavalcanti VO, et al. Characterization of Vibrio cholerae isolated from the aquatic basins of the State of Pernambuco, Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:272–276. doi: 10.1016/j.trstmh.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Mendes CL, Abath FGC, Leal NC. Development of a multiplex single-tube nested PCR (MSTNPCR) assay for Vibrio cholerae O1 detection. Journal of Microbiological Methods. 2008;72(2):191–196. doi: 10.1016/j.mimet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Barrett TJ, Blake PA, Morris GK, Puhr ND, Bradford HB, Wells JG. Use of Moore swabs for isolating Vibrio cholerae from sewage. Journal of Clinical Microbiology. 1980;11:385–388. doi: 10.1128/jcm.11.4.385-388.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pessoa GVA, Da Silva EAM. Medium of Rugai and lysine motility combined in one single tube for the presumptive identification of enterobacteriaceae. Revista do Instituto Adolfo Lutz. 1972;32(1):97–100. [Google Scholar]
- 11.NCID—National Center for Infectious Diseases. Laboratory Identification of V. Cholerae. chapter 6. Atlanta, Ga, USA: CDC/NCID; 1994. Laboratory methods for the diagnosis of Vibrio cholerae ; p. p. 136. http://www.cdc.gov/cholera/pdf/laboratory-methods-for-the-diagnosis-of-vibrio-cholerae-chapter-6.pdf. [Google Scholar]
- 12.Keim P, Price LB, Klevytska AM, et al. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis . Journal of Bacteriology. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. The Lancet. 1993;341(8861):p. 1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 14.Leal NC, Sobreira M, Leal-Balbino TC, et al. Evaluation of a RAPD-based typing scheme in a molecular epidemiology study of Vibrio cholerae O1, Brazil. Journal of Applied Microbiology. 2004;96:447–454. doi: 10.1111/j.1365-2672.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty S, Mukhopadhyay AK, Bhadra RK, et al. Virulence genes in environmental strains of Vibrio cholerae . Applied and Environmental Microbiology. 2000;66:4022–4028. doi: 10.1128/aem.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam MS, Ahsan S, Khan SI, et al. Virulence properties of rough and smooth strains of Vibrio cholerae O1. Microbiology and Immunology. 2004;48(4):229–235. doi: 10.1111/j.1348-0421.2004.tb03518.x. [DOI] [PubMed] [Google Scholar]
- 17.Chun J, Huq A, Colwell RR. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus . Applied and Environmental Microbiology. 1999;65:2202–2208. doi: 10.1128/aem.65.5.2202-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper KLF, Luey CKY, Bird M, et al. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae . Foodborne Pathogens and Disease. 2006;3(1):51–58. doi: 10.1089/fpd.2006.3.51. [DOI] [PubMed] [Google Scholar]
- 19.Rohlf FJ. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System. version 2.11X. Setauket, NY, USA: Exeter Software; 2000. [Google Scholar]
- 20.Dice LR. Measures of the amount of ecological association between species. Ecology. 1945;26:379–393. [Google Scholar]
- 21.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brasil—Ministério da Saúde. Manual Integrado Da Vigilância Epidemiológica Da Cólera. 2008. http://portal.saude.gov.br/portal/arquivos/pdf/manual_integrado_vig_colera.pdf. [Google Scholar]
- 23.Reidl J, Klose KE. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiology Reviews. 2002;26(2):125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 24.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Microbiology: chitin induces natural competence in Vibrio cholerae . Science. 2005;310(5755):1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]


