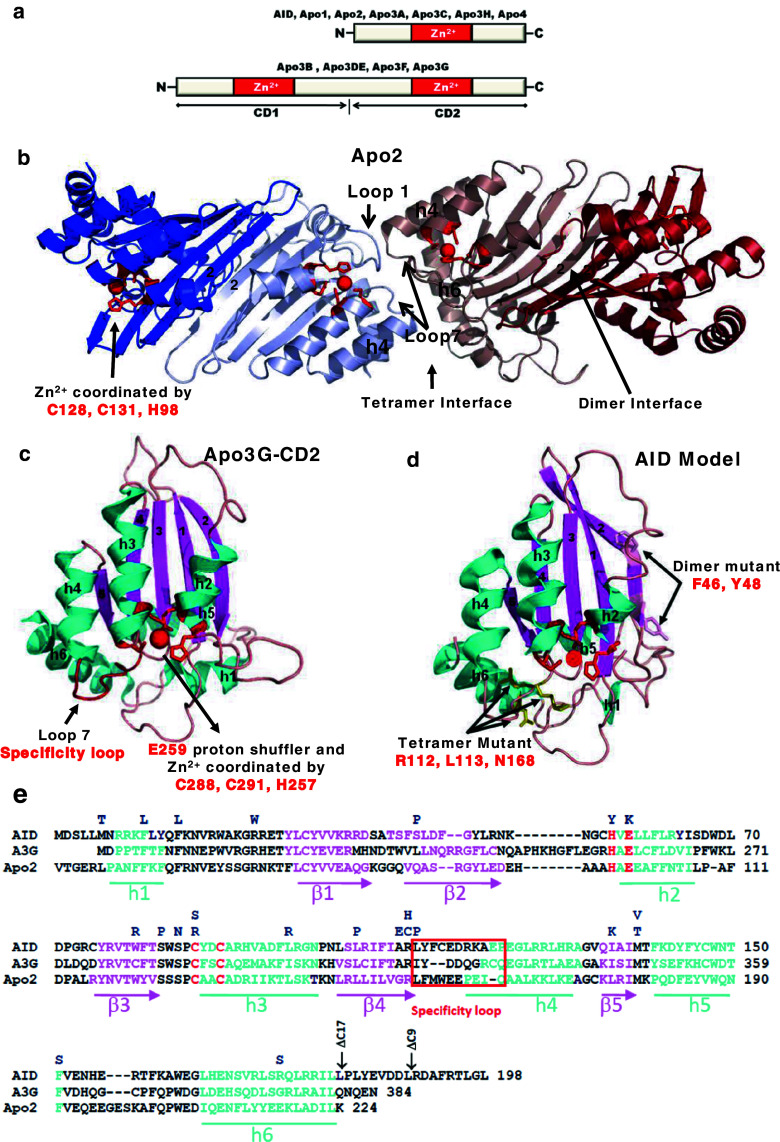
Fig. 3.
Apobec protein domains, structures and sequence alignments. a AID and six other Apobec’s contain a single Zn2+ coordinating deoxycytidine/cytidine deaminase domain, whereas four other members are double-domain deaminases. The N-terminal domain (CD1) of Apo3G, Apo3F, and Apo3DE is inactive while the C-terminal domain (CD2) is catalytically active. b The crystal structure of Apo2 tetramer (Protein Data Bank 2NYT). Head-to-head interaction of two Apo2 dimers is mediated by tetramer interface amino acid residues of α-helix 6, active center loop1, and flexible loop 7. The Apo2 dimer is formed by combining two β2 strands to make one wide β-sheet structure (indicated by arrow). Each monomer is highlighted in a different color. The monomer active site residues C128, C131, and H98 (red) coordinate Zn2+ molecule (red). c The crystal structure of the Apo3G-CD2 (Protein Data Bank 3E1U). The α-helixes are marked in blue and β-strands in pink. The C288, C291, and H257 (red) coordinate Zn2+ molecule (red), whereas E259 (red) acts as a proton shuffler during the hydrolytic deamination reaction. d The AID model was generated using Apo3G-CD2 as a template. Active site residues H56, E58, C87, and C90 (red) are important for catalysis. H56, C87, and C90 are involved in coordinating the Zn ion (red sphere) while E58 is involved in proton shuffling. Predicted residues at the dimeric and tetrameric interfaces are indicated by arrows. e Alignment of AID, Apo3G-CD2, and Apo2. HIGM-2 base substitution mutations and C-terminal deletion mutations are indicated on the top. The amino acids from the protein active site necessary for catalysis are marked in red. The specificity loop is marked in red both in the Apo3G-CD2 structure and amino acid alignment