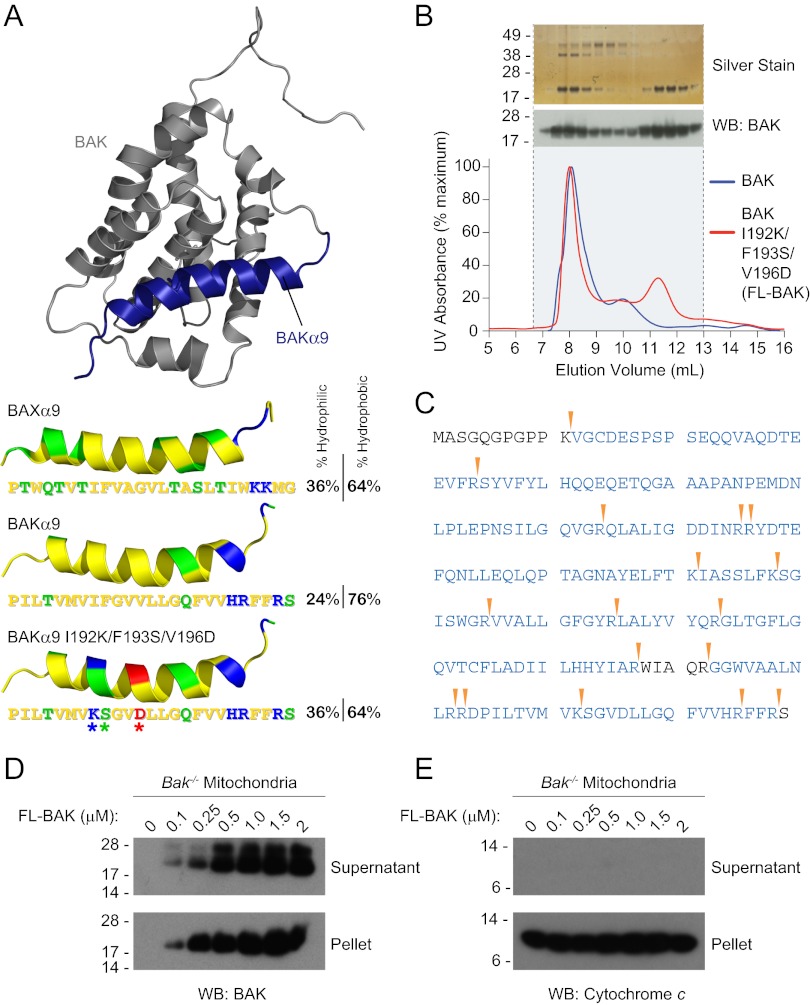
Fig. 1.
Expression and purification of recombinant, full-length, and monomeric BAK. (A) The α9 helix of BAK was subjected to triple mutagenesis (I192K/F193S/V196D) to increase its hydrophilicity and thereby better match that of soluble, full-length BAX. The pictured model structure of BAK was calculated based on the solution structure of BAX using Modeller software (51), with BAK α9 colored blue. The hydrophobic, hydrophilic, positively charged, and negatively charged residues of the BAK and BAX α9 helices are colored yellow, green, blue, and red, respectively. (B) A comparison of the SEC elution profiles of BAK and FL-BAK demonstrates that mutagenesis enabled the isolation of a monomeric species (11- to 12-mL fractions). Silver staining and anti-BAK Western analysis of the electrophoresed FL-BAK fractions documented the isolation of monomeric BAK protein (∼23 kDa). (C) The identity of the isolated monomeric protein was confirmed to be FL-BAK by MS analysis. Tryptic sites are indicated by the arrowheads, and the FL-BAK sequences identified by LC-MS/MS are colored blue. (D) Upon exposure to isolated Bak−/− mouse liver mitochondria, FL-BAK dose-responsively translocated to the mitochondria-containing pellet fraction, as detected by anti-BAK Western analysis. (E) Autotranslocation of FL-BAK to the mitochondrial fraction did not induce cytochrome c release from the pellet to the supernatant, as measured by anti-cytochrome c Western analysis.