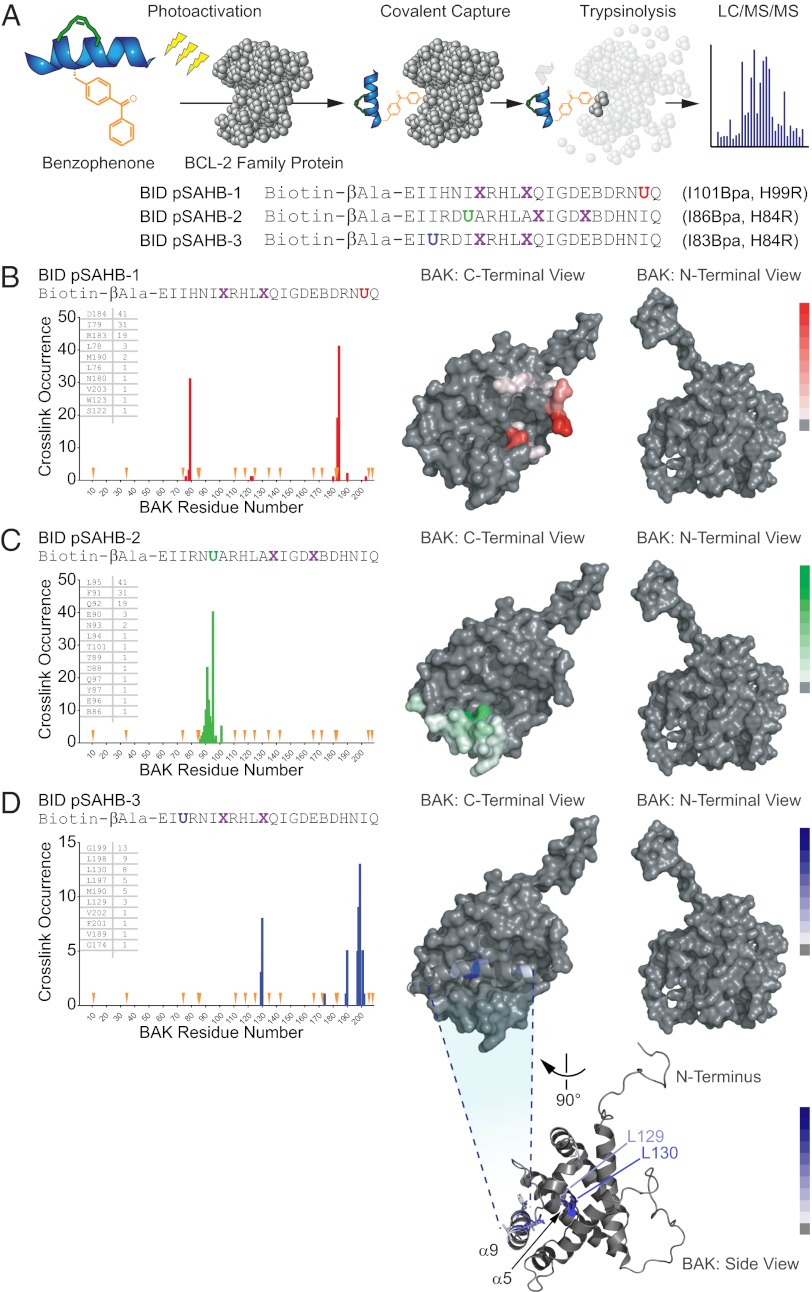
Fig. 4.
Photoreactive BID pSAHBs localize the BH3 trigger site on BAK to its C-terminal BH3-binding pocket. (A) pSAHBs modeled after the BH3 domain of BID were generated for protein capture and binding-site identification by replacing select native residues with 4-benzoyl-L-phenylalanine (Bpa), followed by ring-closing metathesis of olefinic nonnatural amino acids installed at i, i+4 positions. To facilitate tryptic digestion of pSAHBs into shorter and more identifiable fragments by MS, single arginine substitutions were made as indicated, taking advantage of natural sites of homology between the human and mouse BID BH3 domains (i.e., H84R, H99R). X, stapling amino acid; B, norleucine; U, Bpa. (B–D) BID pSAHBs 1–3 were incubated individually with FL-BAK, and the mixtures were subjected to UV irradiation, electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. The plots depict the frequency of crosslinked sites identified across the FL-BAK polypeptide sequence. Crosslinked residues are mapped onto a calculated model structure of FL-BAK (based on sequence homology to BAX) and colored according to the frequency of occurrence for each pSAHB (1, red; 2, green; 3, blue). For BID pSAHBs 1 and 2, which do not crosslink to residues of the C-terminal helix of FL-BAK, α9 has been removed from the structure to better visualize the crosslinked residues at the surface of the canonical BH3-binding pocket. For BID pSAHB-3, a side view of FL-BAK is also shown to demonstrate the localization of crosslinked residues at both the surface of the canonical BH3-binding pocket and the inner surface of α9. The latter dataset shows that, after α9 displacement, previously unexposed and inward-facing α9 residues become available for interaction with BID pSAHB-3.