Fig. 2.
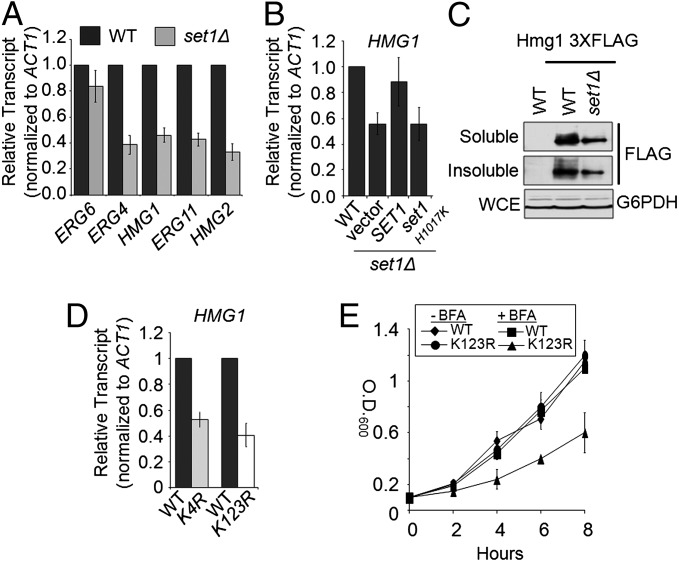
Set1 is necessary for the proper expression of multiple genes in the ergosterol biosynthetic pathway. (A) Expression of ERG6, ERG4, HMG1, HMG2, and ERG11 was determined in the set1Δ strain by qRT-PCR analysis. Statistical analysis determined significant changes in gene expression (P < 0.001) with the exception of ERG6 expression. (B) Expression of HMG1 was determined by qRT-PCR analysis using set1Δ strains transformed with plasmids expressing empty vector, full-length SET1, or catalytically inactive set1 H1017K. Statistical analysis determined significant changes in gene expression (P < 0.001) with the exception of HMG1 expression when full-length Set1 was expressed in the set1Δ strain. (C) Western blot analysis indicating the amount of Hmg1 in WT and set1Δ strains. Glucose 6-phosphate dehydrogenase was used as a loading control. Hmg1 was C-terminally 3xFLAG-tagged at its endogenous locus. (D) Expression of HMG1 was determined by qRT-PCR analysis in the histone H3K4R mutant and H2BK123R mutant strains. All expression analysis was relative to their respective isogenic WT strains using Actin (ACT1) as an internal control to normalize expression levels. Data were analyzed from three biological repeats with three technical repeats. Error bars are SEM. (E) Growth curve of WT and histone H2BK123R mutant strain in the presence or absence of 100 μg/mL BFA.