Figure 1.
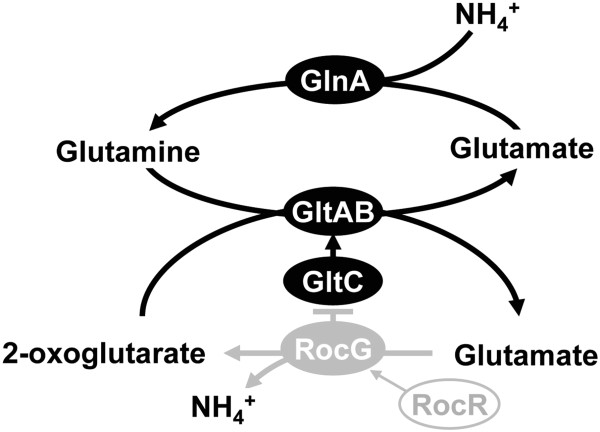
Major reactions and regulation involved in glutamate metabolism in B. subtilis. Proteins are shown as ovals. RocG, glutamate dehydrogenase; GltAB, glutamate synthase (GOGAT); GlnA, glutamine synthetase (GS). In B. subtilis, glutamate can be degraded by RocG. B. subtilis has a glutamine synthetase-glutamate synthase (GS-GOGAT) pathway for assimilation of ammonia. The RocR and GltC transcription factors positively regulate rocG and gltAB, respectively, and GltC can be inhibited via interaction with RocG. Open and closed gray ovals indicate proteins corresponding to genes that have been deleted or inactivated, respectively, in strain MGB874. Deletion of rocR in strain MGB874 decreases expression of rocG, which leads to an increase in expression of gltAB due to activation of GltC via disinhibition by RocG.
