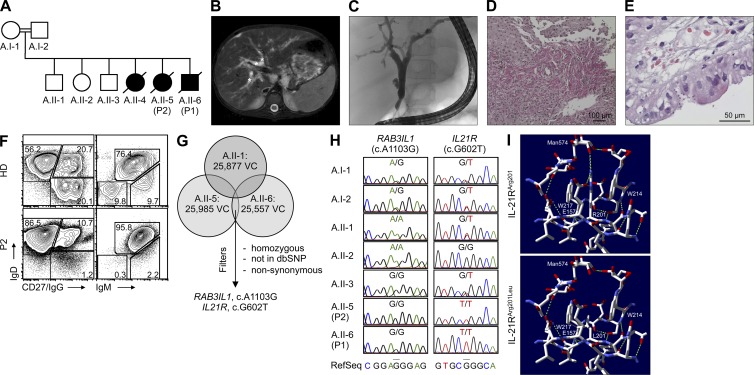
Figure 1.
Clinical and immunological phenotype, identification of IL-21R deficiency, and protein structure analysis in family A. (A) Pedigree of family A. All affected children died secondary to infections and/or therapy-associated complications before the identification of the molecular genetic defect. (B) Abdominal magnetic resonance image revealing hepatomegaly and dilatation of intra- and extrahepatic bile ducts (*) in P2. (C) Endoscopic retrograde cholangiopancreatography demonstrated marked dilatation and irregularities of bile ducts in P2. (D) Histopathological analysis of liver biopsy from P2 showed that fibrous expansion of portal tracts with formation of septa accompanied by prominent ductular proliferation in the portal/lobular interphase was evident. (E) Duodenal biopsy from P2 showed numerous microorganisms ∼2 µm in size and confined to the luminal surface of enterocytes, consistent with cryptosporidiosis. (F) FACS analysis shows accumulation of CD19+IgDhighCD27−IgG− or IgDhighIgMhigh naive B cells in PBMCs isolated from P2 in comparison to a healthy donor. Plots are representative of 3 independent experiments. (G) Scheme of filtering approach for variant calls (VC, single-nucleotide variants and indels [insertion and deletions] <20 bp) from high-throughput sequencing of family members A.II-1, A.II-5, and A.II-6. All variants were filtered according to an autosomal recessive model of inheritance in a consanguineous family. Restricting the candidate set to rare, homozygous, nonsynonymous (NS), or splice site–affecting (SS) variants in the 38 MB CCDS exome resulted in identification of two potentially disease-causing variants: RAB3IL1 and IL21R. (H) DNA Sanger sequencing of the RAB3IL1 and IL21R genes confirmed segregation of the IL-21RArg201Leu mutation with the disease phenotype in family A. (I) Structural analysis of wild-type (Arg201, R201; top) and mutant (Arg201Leu, L201; bottom) IL-21R based on the recently published Protein Data Bank structure 3TGX, illustrating IL-21 complexed with the extracellular domain of the IL-21R (Hamming et al., 2012). Hamming et al. number the amino acids in IL-21R with respect to the first amino acid in chain A of the structure 3TGX, while we number the amino acids in IL-21R in respect to the start of translation. Accordingly, the mutated Arg201 in P1 and P2 refers to Arg182 in (3TGX, Hamming et al., 2012). (top) The neighborhood of wild-type Arg201 displaying putative hydrogen bonds with a sugar chain and the Glu157 reside (green dashed lines). (bottom) The same region in the mutated IL-21R structure, illustrating that the Arg201Leu substitution is predicted to break these putative hydrogen bonds. Steric clashes are shown in purple.