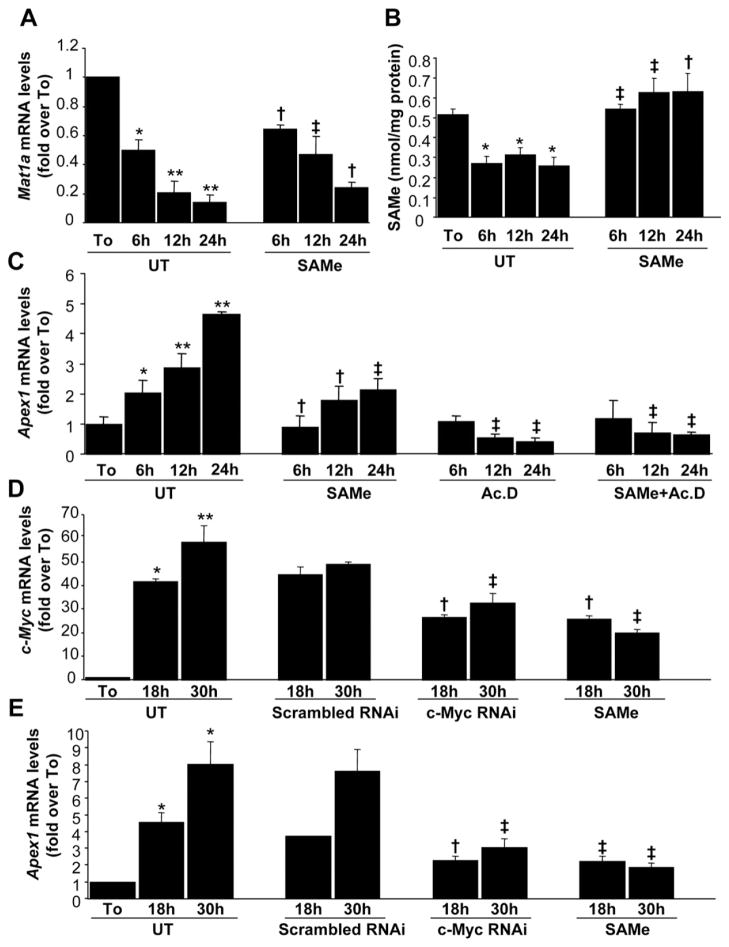
Figure 4.
Effect of SAMe treatment on Mat1a, Apex1, and c-Myc mRNA levels in mouse hepatocytes. Mouse hepatocytes were treated with SAMe (2 mmol/L) for different time points. When indicated, cells were pretreated with actinomycin D (5 μg/mL) for 5 minutes. At the indicated time points, RNA from hepatocyte suspension right after isolation (To) or cultured hepatocytes were extracted for real-time PCR. (A) Effect of SAMe treatment on Mat1a mRNA levels. Results are expressed as fold over To cells (mean ± SEM) from 3 independent experiments. *P < .005 vs To, **P < .0001 vs To, †P < .05 vs untreated (UT) at respective time points, ‡P < .01 vs UT at respective time points. (B) Effect of SAMe treatment on intracellular SAMe levels. Hepatocytes were treated with SAMe (2 mmol/L) for different time points. Cellular SAMe levels were measured by high-performance liquid chromatography. The results are expressed as nanomoles of SAMe per milligrams of protein (mean ± SEM) from 3 independent experiments. *P < .005 vs To, †P < .05 vs UT at respective time points, ‡P < .001 vs UT at respective time points. (C) Effect of SAMe treatment on Apex1 mRNA levels as determined by real-time PCR. Results are expressed as fold over To cells (mean ± SEM) from 3 independent experiments. *P < .04 vs To, **P < .001 vs To, †P < .04 vs UT at respective time points, ‡P < .001 vs UT at respective time points. (D) Effect of c-Myc RNAi and SAMe treatments on c-Myc mRNA levels. Hepatocytes were treated with scrambled or c-Myc RNAi as described in the Materials and Methods section or with 2 mmol/L SAMe for indicated times. RNA was subjected to quantitative real-time PCR analysis. Results are expressed as fold over To cells (mean ± SEM) from 4 independent experiments. *P < .004 vs To, **P < .0001 vs To, †P < .05, ††P < .0001 vs UT at respective time points. (E) Effect of c-Myc RNAi and SAMe treatments on Apex1 mRNA levels. The same samples from D were subjected to real-time PCR for Apex1 mRNA measurement. *P < .0002 vs To, †P < .004, ††P < .002 vs UT at respective time points.