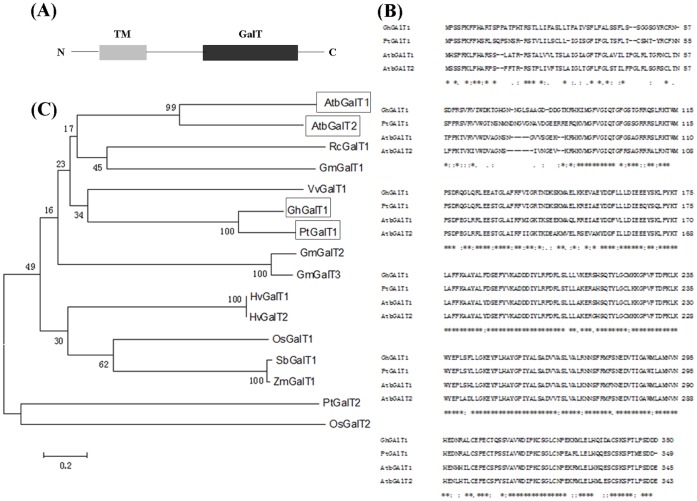
Figure 1. Characterisation of GhGalT1.
(A) Structure of GhGalT1 protein. N: N-terminal; TM: transmembrane domain, GalT: the conservative galactosyltransferase functional domain; C: C-terminal. (B) Comparison of the predicted amino acid sequence of GhGalT1 with some known GalT proteins. The alignment was generated with the CLUSTAL W program. The same nucleotides are highlighted by asterisk, conserved and similar residues are indicated by colon or dot mark. (C) Phylogenetic relationships between GhGalT1 and other GalTs in plants. A neighbour-joining tree was generated with MEGA5 from 1000 bootstrap replicates. The accession numbers of these known proteins in GenBank are: Gossypium hirsutum GhGalT1 (JX448620); Populus trichocarpa PtGalT1 (XP_002330737) and PtGalT2 (XP_002317466); Arabidopsis thaliana AtbGalT1 (At1g53290) and AtbGalT2 (At3g14960); Zea mays ZmGalT1 (NP_001152267); Vitis vinifera VvGalT1 (XP_002283081); Ricinus communis RcGalT1 (XP_002524276); Glycine max GmGalT1 (XP_003543216), GmGalT2 (XP_003556804) and GmGalT3(XP_003527208); Oryza sativa OsGalT1 (Os06g0156900) and OsGalT2(CAD44839.); Hordeum vulgare HvGalT1 (BAJ94890) and HvGalT2(ABL11234); Sorghum bicolor SbGalT1 (XP_002436508).