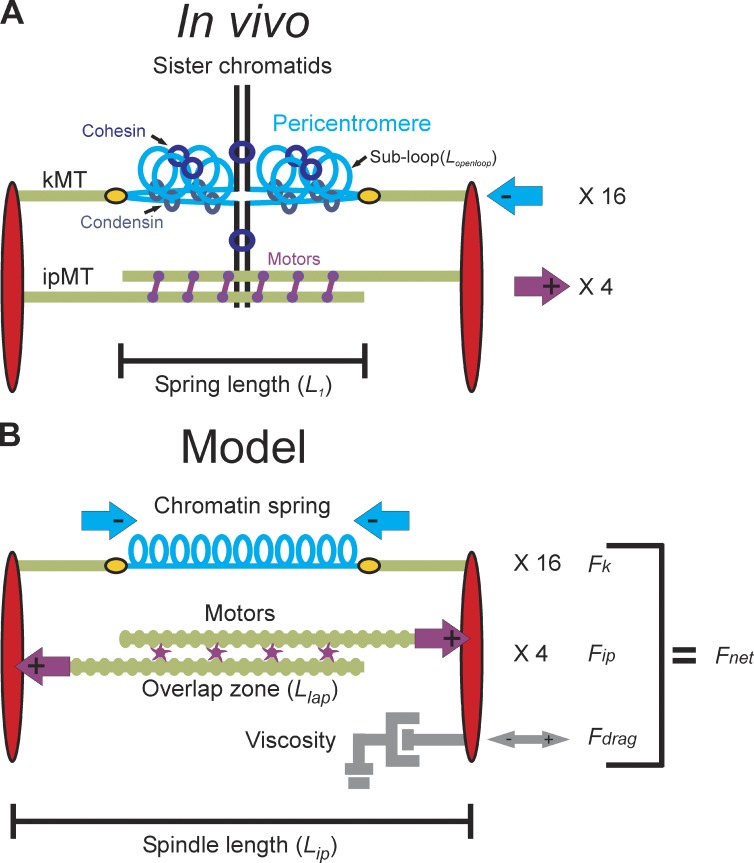
Figure 1.
Structure of the yeast mitotic spindle. (A) Microtubules (kMTs, green) emanating from opposite spindle pole bodies (red) bind to the centromere via the kinetochore (yellow). Sister centromeres are spatially separated in metaphase and reside at the apex of a pericentric chromatin loop (in blue) that extends perpendicularly from the chromosome axis (black; Yeh et al., 2008). The total contour length of the pericentric chromatin loop is split between an axial component (approximated by the distance between the two kinetochores, L1) and subloops (Lopenloop) that extend perpendicular to the spindle axis. Condensin is more proximal to the spindle axis than cohesin. Approximately eight ipMTs overlap (two shown) and are bound by kinesin 5 motor proteins (purple). Although only one replicated chromosome is depicted with two kMTs, there are 16 chromosomes (32 sister chromatids) in budding yeast and ∼32 kMTs. The 16 kinetochores from each pole are clustered in mitosis. The aggregate chromatin spring length is measured by the distance between the two clusters (L1). (B) The model is written as a coupled system of stochastic and deterministic differential equations in which the sum of the forces applied to one spindle pole body is used to numerically solve for velocity at each time step. Spindle length is defined experimentally as the distance between the spindle pole bodies (red) in metaphase (Lip). The pericentric chromatin functions as a spring (blue, Fk). Its length is the distance between two sister kMT plus ends (Lspring). Kinesin motors (purple) bind to and couple ipMTs at the overlap zone (Llap) and slide ipMTs apart, generating an outward extensional force, Fip. The viscous properties of the nucleus are represented as a dashpot and resist movement of the spindle pole bodies in either direction (gray, Fdrag).