Processing of unusual replication intermediates such as reversed forks by MUS81 contributes to oncogene-induced double-strand breaks and depends on mitotic entry.
Abstract
Oncogene-induced DNA replication stress activates the DNA damage response (DDR), a crucial anticancer barrier. DDR inactivation in these conditions promotes genome instability and tumor progression, but the underlying molecular mechanisms are elusive. We found that overexpression of both Cyclin E and Cdc25A rapidly slowed down replication forks and induced fork reversal, suggestive of increased topological stress. Surprisingly, these phenotypes, per se, are neither associated with chromosomal breakage nor with significant DDR activation. Oncogene-induced DNA breakage and DDR activation instead occurred upon persistent G2/M arrest or, in a checkpoint-defective context, upon premature CDK1 activation. Depletion of MUS81, a cell cycle–regulated nuclease, markedly limited chromosomal breakage and led to further accumulation of reversed forks. We propose that nucleolytic processing of unusual replication intermediates mediates oncogene-induced genotoxicity and that limiting such processing to mitosis is a central anti-tumorigenic function of the DNA damage checkpoints.
Introduction
Activation of a growing number of oncogenes has been found associated with “replication stress,” a poorly understood perturbation of DNA replication (Mailand et al., 2000; Bartkova et al., 2005, 2006; Gorgoulis et al., 2005; Di Micco et al., 2006; Dominguez-Sola et al., 2007). Replication stress induces the activation of the DNA damage response (DDR), which is detected from the earliest stages of tumorigenesis (Bartek et al., 2007). Oncogene-induced replication stress is associated with the formation of double-strand breaks (DSBs), particularly in regions intrinsically difficult to replicate (Bartkova et al., 2005, 2006; Gorgoulis et al., 2005). The observed DDR activation induces senescence in precancerous lesions and functions as a barrier against full malignant transformation (Bartkova et al., 2006; Di Micco et al., 2006).
Oncogene activation affects—directly or via deregulation of CDK2—the replication initiation program, resulting in deregulated origin firing and impaired fork progression. The latter effect is proposed to result from nucleotide depletion (Bester et al., 2011), from interference between DNA replication and transcription (Jones et al., 2012), and/or from increased DNA torsional stress (Bermejo et al., 2012), but the lack of structural information on replication intermediates (RIs) under these experimental conditions has so far limited our understanding of the underlying mechanisms. Furthermore, it is unclear how perturbations at the replication forks lead to DSB formation that promotes chromosomal rearrangements during tumorigenesis.
Replication stress has been recently associated with transient accumulation of DNA lesions and large 53BP1 foci formed when cells progress through mitosis (Lukas et al., 2011). It was previously reported that different oncogenes lead to mitotic aberrations (Molinari et al., 2000; Ichijima et al., 2010), but the causative link between these phenotypes and oncogene-induced genotoxic stress has remained obscure. Recently, additional molecular events of potential importance for chromosomal integrity were associated with mitotic entry, e.g., the resolution of Holliday junctions (HJs), central intermediates of DNA homologous recombination (HR; Matos et al., 2011; Schwartz and Heyer, 2011). Furthermore, the HJ resolvase MUS81 was recently implicated in DSB formation upon oncogene overexpression (OE) or cell cycle perturbation, but the link to mitotic progression is controversial and the underlying mechanisms remained elusive (Beck et al., 2010; Domínguez-Kelly et al., 2011; Forment et al., 2011; Murfuni et al., 2013).
In this work, comparing OE of Cyclin E (CycE) and Cdc25A, we identify reversed forks as common unusual replication intermediates, suggesting increased topological stress as a critical determinant of oncogene-induced replication stress. Surprisingly, fork slowing and restructuring are, per se, neither associated with chromosomal breakage nor with full DDR activation. We show that processing of these unusual replication intermediates by MUS81 depends on mitotic entry and contributes to oncogene-induced DSBs. Premature CDK1 activation upon checkpoint inactivation accelerates and exacerbates oncogene-induced DSB formation, providing a mechanistic rationale for the function of the DNA damage checkpoints as barriers against genome instability. Thus, specific DNA structures and cell cycle transitions mediate oncogene-induced chromosomal breakage.
Results and discussion
Oncogene OE rapidly interferes with replication fork progression and induces fork reversal
To elucidate the impact of oncogenes on the replication process, we focused on two established systems of oncogene OE (Mailand et al., 2000; Bartkova et al., 2005) as prototypes of two different scenarios: (1) “Oncogene OE only” by CycE OE, where CDK2 hyperactivation deregulates DNA replication; and (2) “Oncogene OE + checkpoint defect” by Cdc25A OE. As this phosphatase is at the same time a CDK activator and a key effector of the DNA damage response (Mailand et al., 2000), Cdc25A OE combines CDK2 deregulation with impaired cell cycle control, two key steps in tumorigenesis (Bartkova et al., 2006; Di Micco et al., 2006).
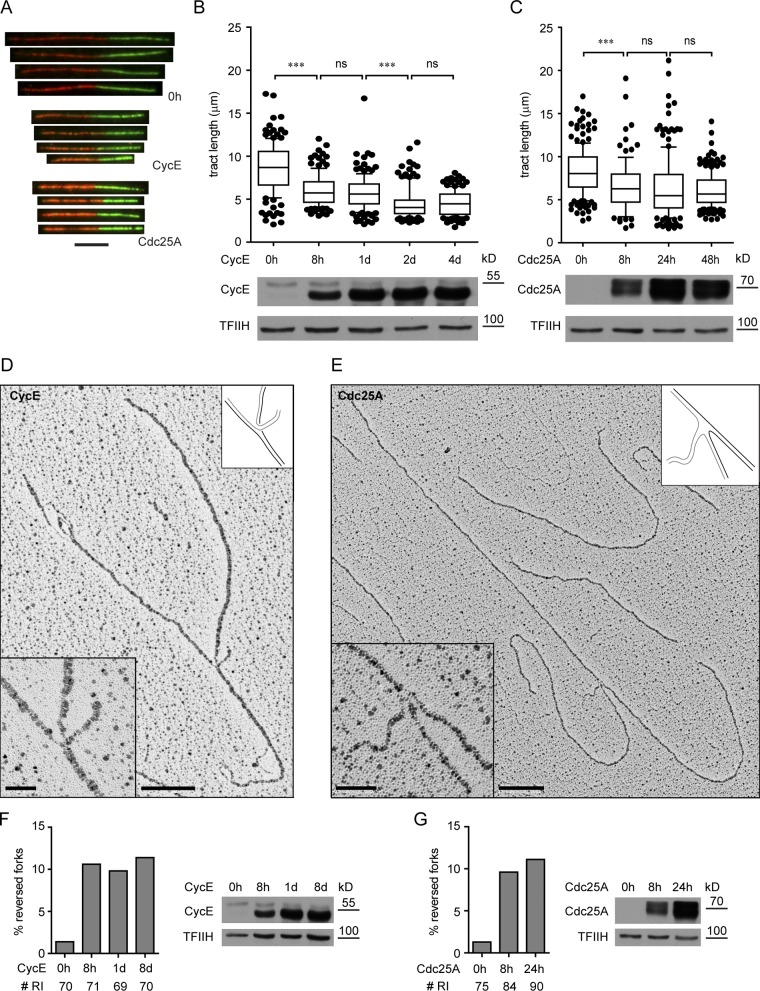
We tested the effect of both oncogenes on the progression of individual replication forks by DNA fiber spreading analysis (Fig. 1 A; Jackson and Pombo, 1998). In keeping with two recent reports (Bester et al., 2011; Jones et al., 2012), oncogene OE is associated with a significant replication fork slowdown. We now show that this fork delay does not require prolonged oncogene expression (Bester et al., 2011), but is detectable in both systems from the earliest time point (8 h; Fig. 1, A–C). Next, we investigated in vivo replication fork architecture upon oncogene OE by electron microscopy (EM; Neelsen et al., 2013). Our most striking observation was the accumulation of reversed forks (RFs), i.e., replication forks showing a fourth regressed arm, due to local annealing of the newly synthesized strands (Fig. 1, D and E). Although these are rare intermediates during unperturbed S phase, they rapidly accumulate upon OE of both CycE and Cdc25A (Fig. 1, D–G; and Fig. S1 A). Furthermore, their frequency does not further increase at later time points (Fig. 1, F and G). Upon OE of both oncogenes, we also detected an increasing number of molecules exposing extended single-stranded DNA (ssDNA) regions, mainly as gaps on newly synthesized strands, but also at the replication fork (Fig. S1, B–F). In Cdc25A-overexpressing cells, a few replication bubbles showed one side entirely single stranded (hemireplicated bubble; Fig. S1 C; Sogo et al., 2002). Moreover, in a subset of RIs from later time points of CycE OE, we occasionally detected ssDNA gaps on the parental strands (Fig. S1 D), suggesting that ssDNA regions are carried over from the previous round of replication. To exclude that our results are specific for cancer cells, we transiently overexpressed both oncogenes in untransformed human MRC5 fibroblasts. Transient CycE and Cdc25A OE in MRC5 resulted in replication fork slowdown similar to that in U2OS cells, and was accompanied by the accumulation of RF- and ssDNA-containing molecules (Fig. S2, A–D). Overall, these data identify common features of DNA replication stress—i.e., fork slowing, fork reversal, and ssDNA accumulation—detectable with similar kinetics upon OE of both oncogenes, and provide direct evidence that oncogene OE affects the structure of replication intermediates in vivo.
Figure 1.
Oncogene OE slows down replication forks and induces fork reversal. (A) DNA tracts pulse-labeled with CldU and IdU from control cells (0 h) and cells overexpressing the indicated oncogene for 8 h. (B and C) Analysis of replication fork progression by DNA fiber spreading before (0 h) and after induction of CycE, and Cdc25A, respectively. Bottom panels show oncogene OE. (D and E) Electron micrographs of reversed forks from cells overexpressing CycE and Cdc25A, respectively. Insets show magnified forks and schemes of fork structure. Black and gray lines describe parental and newly synthesized DNA strands, respectively. (F and G) Frequency of reversed replication forks after induction of CycE and Cdc25A, respectively. “# RI” is the number of analyzed replication intermediates. Panels on the right show oncogene OE. Data in F and G were reproduced in at least one independent experiment. At least 100 tracts were scored per sample in B and C. Whiskers: 10–90th percentile (***, P < 0.0001; ns, not significant, Mann-Whitney test). Bars: (A) 5 µm; (D and E) 200 nm (500 bp); (insets) 50 nm. TFIIH as loading control. Molecular weight in kD of nearest protein size marker is indicated.
Cell cycle progression, chromosomal breakage, and DDR upon CycE and Cdc25A OE
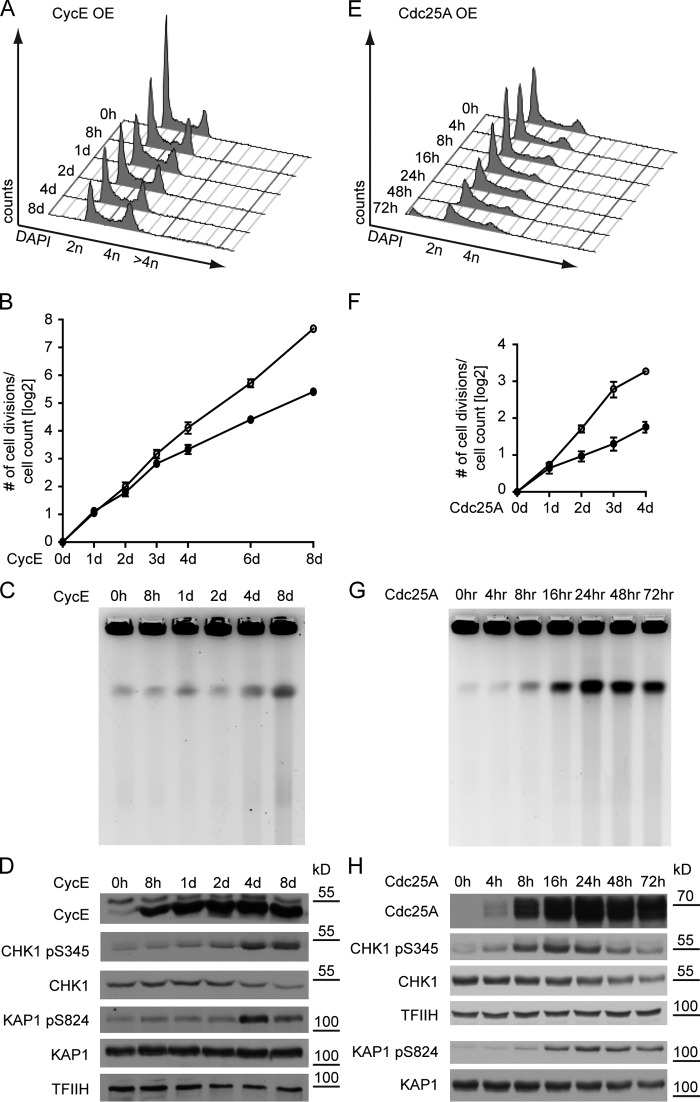
We next tested whether replication stress is associated with DSBs, cell cycle arrest, and DDR activation. CycE OE leads to transient accumulation of cells in S and G2/M phases, followed at late time points (4–8 d) by accumulation of rereplicating cells (DNA content >4n; Fig. 2 A). Cell cycle deregulation by CycE OE delayed proliferation only three days after oncogene induction (Fig. 2 B). Mild ATR/CHK1 activation was detectable within 24 h of CycE OE, whereas DNA breakage—as assessed by pulse-field gel electrophoresis (PFGE) —and ATM/KAP1 phosphorylation were detected only after 4–8 d (Fig. 2, C and D), when the cells accumulate in G2/M and occasionally rereplicate (Fig. 2 A). Thus, in this experimental system, oncogene-induced fork slowing and reversal precede and are not directly associated with DSBs (Fig. 1, F and G; and Fig. 2). In contrast, Cdc25A OE leads to a marked arrest of cell cycle and proliferation already one day after induction (Fig. 2, E and F). ATR/CHK1 activation was detectable early after oncogene OE (8 h) and was rapidly associated with massive DNA breakage (16–24 h) and ATM/KAP1 activation (Fig. 2, G and H).
Figure 2.
Oncogene OE deregulates the cell cycle and causes DNA breakage and bi-phasic DDR activation. (A and E) FACS analysis of DNA content (DAPI) of cells overexpressing CycE for up to 8 d and Cdc25A for up to 72 h, respectively. For additional data on the accumulation of cells with >4n DNA, see Figs. 3 B and S3 H. Data are from a single representative experiment out of four repeats. (B and F) Proliferation of cells overexpressing CycE and Cdc25A, respectively (mean + SEM, n = 4). (C and G) DNA breakage after CycE and Cdc25A OE monitored by pulse-field gel electrophoresis. (D and H) DDR activation (pCHK1, pKAP1) and total DDR proteins (CHK1, KAP1) upon OE of CycE and Cdc25A, respectively. TFIIH serves as loading control for D and the top panels of H; total KAP1 levels control for KAP1 pS284 in the bottom panels of H. Molecular weight in kD of nearest protein size marker is indicated.
Transient OE of CycE and Cdc25A in MRC5 yielded similar results, in that CycE OE for 48 h neither caused DSB nor DDR activation, whereas Cdc25A OE resulted in DSB formation and stepwise activation of ATR/CHK1 and ATM/KAP1 (Fig. S2, E and F). Overall, CDK2 deregulation by both CycE and Cdc25A OE rapidly induces prolonged S phase and ATR activation, and accumulation of unusual replication intermediates. However, chromosomal breakage and cell cycle arrest occur with strikingly different kinetics in the two systems.
Marked replication stress does not activate the DDR until cells experience a persistent G2/M arrest (CycE) or a premature replication block (Cdc25A)
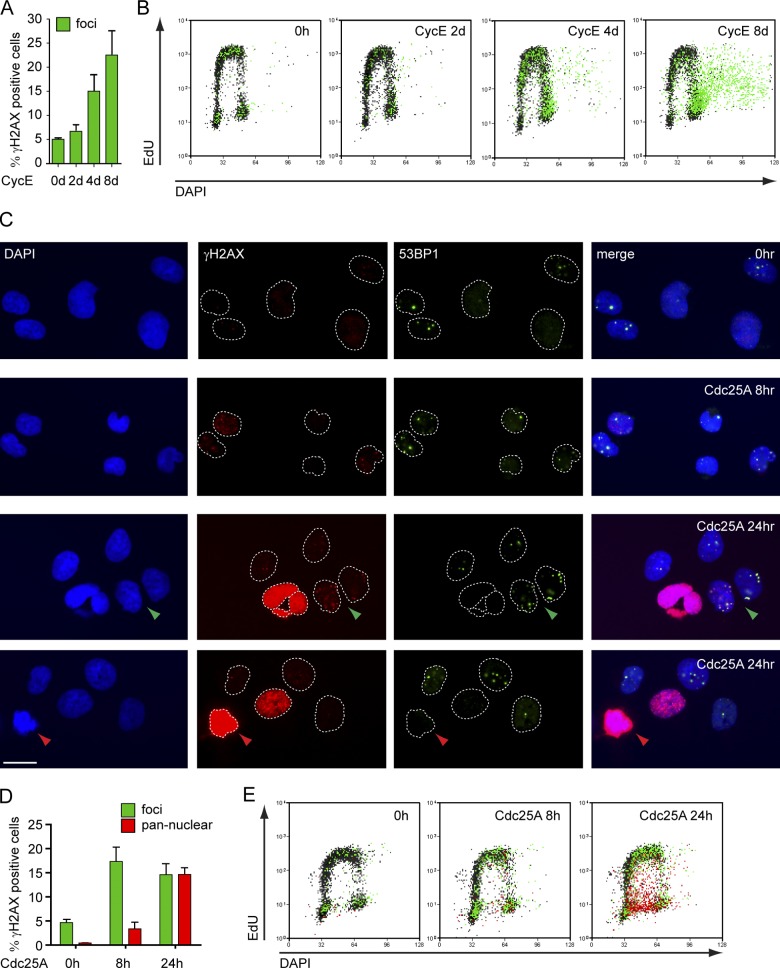
To further characterize oncogene-induced DNA breakage and DDR, we studied H2AX phosphorylation (γ-H2AX) and 53BP1 recruitment by single-cell immunofluorescence (IF). Whereas the former event marks sites of DNA damage in general, the latter is more specific for DSBs (de Feraudy et al., 2010; Ray Chaudhuri et al., 2012). Despite the marked replication phenotypes (Figs. 1 and S2), CycE-overexpressing cells showed no γ-H2AX above background for 2–3 d after induction. 10–20% of the cells did show γ-H2AX foci at later time points (4–8 d), when DSBs become physically detectable (Figs. 2 C, 3 A, and S3 A). Coupling IF-based γ-H2AX detection with flow-cytometric analysis of DNA content (DAPI) and replication (5-ethynyl-2′-deoxyuridine [EdU]; Fig. S3, B and C), we observed that increased γ-H2AX levels upon prolonged CycE OE are mainly present in cells accumulating in G2/M or attempting rereplication (Fig. 3 B). Rereplicating cells were also easily identified by IF microscopy because of their “giant nuclei” (Zhu et al., 2004) and displayed accumulation of γ-H2AX foci, mostly colocalizing with 53BP1, thus marking DSBs (Fig. S3 D). In contrast, the DDR observed upon Cdc25A OE was more rapid and heterogeneous. Cells with γ-H2AX/53BP1 foci were detected 8 h after Cdc25A induction, whereas after 24 h a significant fraction of cells showed intense pan-nuclear γ-H2AX (Fig. 3, C and D), a phenotype previously associated with replication stress (Murga et al., 2009). Pan-nuclear γ-H2AX was consistently associated with intermediate DNA content and compromised EdU incorporation, suggesting a replicative arrest (Fig. 3 E). These cells also consistently show intense DAPI staining and, in spite of the high γ-H2AX signal, are devoid of 53BP1 foci (Fig. 3 C). Both features are characteristic of mitotic chromatin condensation (Giunta et al., 2010). Cdc25A OE also rapidly induced an increase in nuclear fragmentation, frequently associated with pan-nuclear γ-H2AX (Fig. 3 C; Fig. S3, E and F). A similar phenotype was associated with prolonged CycE OE (Fig. S2 G). In summary, CycE-overexpressing cells show mild and slow accumulation of DSB markers in IF and FACS. In contrast, these markers are rapidly detectable upon Cdc25A OE and correlate with mitotic features in cells experiencing a replicative arrest.
Figure 3.
Different kinetics, extent, and cell cycle dependency of DDR activation upon Cdc25A and CycE OE. (A) γ-H2AX–positive cells (green) before (0 h) and at the indicated time points after CycE induction quantified by single-cell IF (see also Fig. S3). (B) FACS analysis of DNA synthesis (EdU), DNA content (DAPI), and DDR activation (γ-H2AX) in cells overexpressing CycE. γ-H2AX–positive cells (foci) in green. (C) Single-cell IF of control cells (0 h) and cells overexpressing Cdc25A for 8 h and 24 h stained for γ-H2AX and 53BP1. Green arrowhead, cell with γ-H2AX foci. Red arrowhead, cell with pan-nuclear γ-H2AX. (D) γ-H2AX–positive cells quantified by single-cell IF, and (E) FACS analysis of cells overexpressing Cdc25A. Cells with γ-H2AX foci in green and pan-nuclear γ-H2AX in red. (A and D) mean + SEM, n = 4. Bar, 10 µm. For OE data on CycE and Cdc25A, see Fig. S3. Data in B and E are from a single representative experiment out of four repeats.
Massive chromosomal breakage and DDR are associated with premature activation of mitotic markers and suppressed by CDK1 inhibition
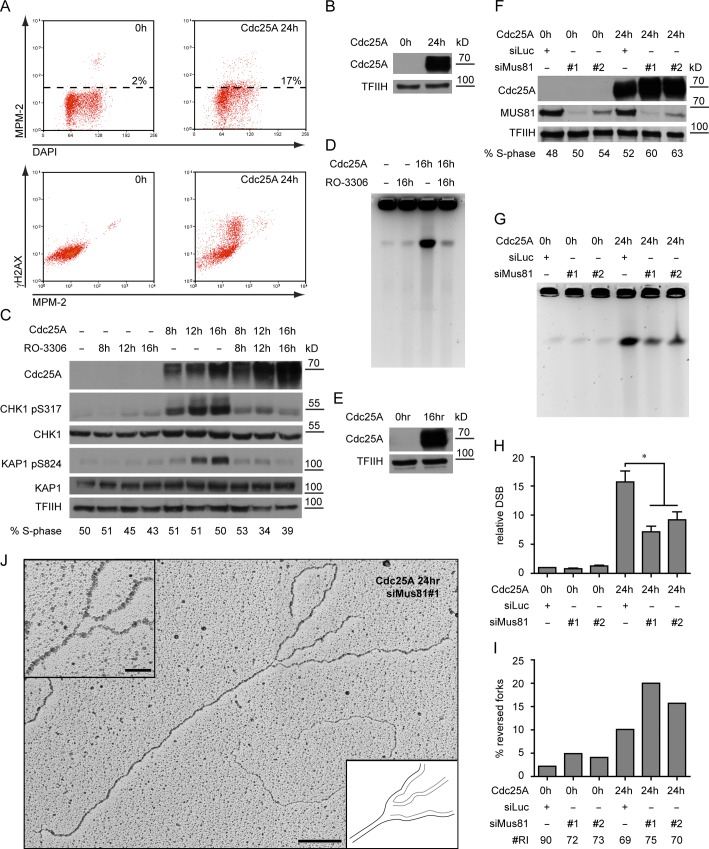
The results shown so far imply that oncogene-induced replication stress does not directly lead to DSB formation. However, they suggest that oncogene-induced DSBs occur either in mitosis or upon premature mitotic entry. To test this hypothesis, we used a broad-spectrum marker of mitotic CDK1-activity—the MPM-2 antibody, which recognizes an abundant phospho-epitope on CDK1 substrates (Davis et al., 1983)—and found that 24 h after Cdc25A induction this marker is no longer restricted to mitotic cells, but detectable also in a substantial fraction of cells with intermediate DNA content (Fig. 4, A and B). Furthermore, intense γ-H2AX in Cdc25A-overexpressing cells is associated with elevated levels of MPM-2 (Fig. 4 A), establishing a link between premature mitotic entry and oncogene-induced DDR activation. To assess directly whether initiation of mitosis is required for Cdc25A-induced DNA breakage, we overexpressed the oncogene in the presence of the CDK1 inhibitor RO-3306 (Vassilev et al., 2006). CDK1 inhibition did not interfere with oncogene expression, allowed transit into S phase, and did not affect the initial (8 h) increase of CHK1 activity induced by Cdc25A OE (Figs. 4 C and S3 I). However, Cdc25A-induced DSB and ATM activation were completely suppressed (Fig. 4, C–E). Accordingly, CHK1 phosphorylation after 12–16 h, presumably resulting from DSB resection, was also suppressed (Fig. 4 C). In checkpoint-proficient cells, the ATR pathway restricts CDK1 activity in response to replication stress. To test whether ATR limits oncogene-induced DNA breakage, we inhibited ATR in CycE-overexpressing cells (Toledo et al., 2011). Indeed, ATR inhibition increased the amount of CycE-induced DSBs after prolonged CycE OE (Fig. S3, J–L). Taken together, our data indicate that the massive chromosomal breakage induced by Cdc25A OE is associated with and depends on premature initiation of mitosis.
Figure 4.
Cdc25A-induced DSBs depend on CDK1-mediated mitotic entry and on MUS81-dependent processing of reversed forks. (A) FACS analysis for phosphorylation of CDK1 substrates (MPM-2) and DDR activation (γ-H2AX) of control cells (0 h) and cells overexpressing Cdc25A for 24 h. High levels of γ-H2AX in unperturbed mitotic cells have been previously reported (McManus and Hendzel, 2005). Data are from a single representative experiment out of three repeats. (B) Cdc25A OE in samples in A. (C) Activation of the DDR (pCHK1, pKAP1) and total DDR proteins (CHK1, KAP1) upon Cdc25A OE of Cdc25A in the absence or presence of the CDK1 inhibitor. S phase scored by EdU incorporation. (D) Cdc25A-induced DNA breakage assessed by PFGE, in the absence or presence of the CDK1 inhibitor. (E) Cdc25A OE in samples in D. (F) Immunoblot showing OE of Cdc25A and depletion of MUS81. S phase scored by EdU incorporation. (G) DNA breakage monitored by pulse-field gel electrophoresis before (0 h) and 24 h after induction of Cdc25A in mock- or MUS81-depleted cells. (H) Quantification of chromosomal breakage by PFGE in cells treated as in (F), mean + s.e.m., n ≥ 3, * = P < 0.05, Paired student’s t test. (I) Frequency of reversed replication forks in cells treated as in F. “# RI” is the number of analyzed replication intermediates. (J) Micrograph of a reversed replication fork from MUS81-depleted cells overexpressing Cdc25A. The regressed arm is connected to one of the daughter strands, leaving a gap at the branch point. Data in I was reproduced in one independent experiment. Bar: (main panel) 200 nm (500 bp); (inset) 50 nm. TFIIH as loading control. Molecular weight in kD of nearest protein size marker is indicated. For FACS profiles quantified in C and F, see Fig. S3.
Reversed forks are MUS81 substrates and precursors of oncogene-induced DSBs
The HJ resolvase MUS81-EME1 was previously suggested to process replication forks after prolonged arrest (Hanada et al., 2006, 2007), and was also recently implicated in oncogene-induced genotoxicity (Murfuni et al., 2013), but neither its mechanistic role nor its substrates have been identified. We found that siRNA-mediated MUS81 depletion suppressed up to 60% of Cdc25A-induced breaks (Fig. 4, F–H; and Fig. S3 M). As we identified RFs as an abundant unusual intermediate upon oncogene OE, we tested the hypothesis that they are substrates for cleavage by MUS81. Indeed, MUS81 depletion caused an increase in Cdc25A-induced RFs, strictly correlating with the amount of residual protein and the decrease of DSBs (Fig. 4, F–I). A reproducible increase in RF frequency was also observed in the absence of oncogene OE, suggesting that occasional RFs formed in unperturbed conditions are targeted by MUS81. Overall these data show that RFs are substrates for MUS81 cleavage and precursors of oncogene-induced DNA breakage.
Our work sheds light on several important mechanistic aspects of oncogene-induced genotoxicity. We present the first direct visualization of the impact of oncogene OE on the structure of RIs in vivo. Although the observed accumulation of ssDNA was predicted by previous studies (Bartkova et al., 2005), it is surprising that replication forks challenged by OE of both CycE and Cdc25A regress rapidly and frequently. Reversed forks were first shown in bacteria in response to torsional stress (Postow et al., 2001). They had been long postulated also in eukaryotic cells (Higgins et al., 1976) and were shown to arise in yeast upon topological impediments induced by checkpoint defects (Sogo et al., 2002; Bermejo et al., 2011). Most recently, reversed forks have been reported as frequent RI upon topoisomerase I poisoning also in higher eukaryotes (Ray Chaudhuri et al., 2012). Overall, the available data establish fork reversal as an evolutionary conserved response to topological constraints. Our structural observations support a scenario where oncogene OE impairs the replication process by inducing topological stress resulting from deregulating simultaneously CDK2-dependent origin firing and transcription (Fig. 5; Bermejo et al., 2012). In line with this notion, most oncogenes shown to induce replication stress deregulate the G1–S transition (Bartkova et al., 2005, 2006; Di Micco et al., 2006). Accordingly, CycE-induced fork slowing was recently linked to high levels of replication initiation and the resulting interference between replication and transcription (Jones et al., 2012). Prolonged exposure to oncogenic stress was also recently linked to nucleotide depletion (Bester et al., 2011), which may arise as a consequence of supernumerary replication factories and contribute to replication fork stalling.
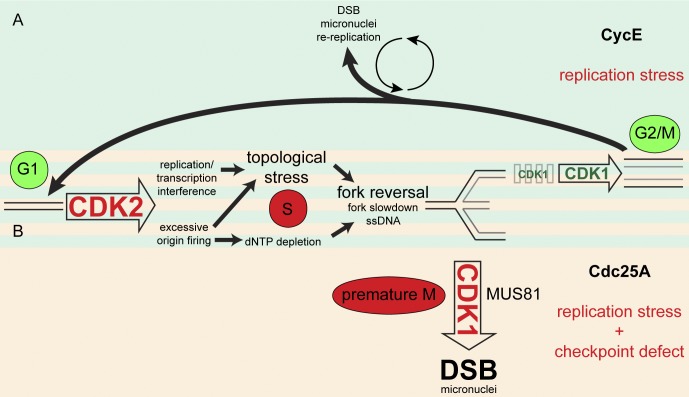
Figure 5.
Replication stress and DNA damage induction upon CycE and Cdc25A OE. (A) As many other oncogenes (e.g., Cdc25A), CycE OE causes CDK2 hyperactivation, deregulating the replication program and leading to topological stress. This impairs fork progression and promotes fork reversal. In the following G2/M, residual replication intermediates can be resolved into transient DNA lesions (Lukas et al., 2011), which are repaired in G1. Accumulation of replication stress upon prolonged oncogene OE leads to persistent G2/M arrest, mitotic defects (micronuclei), and rereplication associated with low DSB levels. (B) When CDK2 deregulation is associated with DNA damage checkpoint defects, as upon Cdc25A OE or in later stages of tumorigenesis, cells are no longer able to delay CDK1 activation. Premature CDK1 activation leads to mitotic aberrations (micronuclei) and to massive DNA breakage due to MUS81-mediated processing of reversed forks. Physiological and oncogene-induced pathological events are indicated in green and red, respectively. Further details are discussed in the text.
A second unexpected conclusion of our work is that the manifestations of oncogene-induced replication stress (fork reversal and ssDNA exposure) are not, per se, leading to chromosomal breakage, but are tolerated without potent DDR activation for at least three cell cycles upon CycE OE. In contrast, Cdc25A-overexpressing cells show massive DNA breakage, pan-nuclear γ-H2AX, and cell cycle arrest already 16–24 h after induction. Our data identify the critical determinant of this difference in the regulation of CDK1-dependent mitotic entry, which is maintained upon oncogene OE (CycE OE), but lost with severe cellular consequences upon inactivation of cell cycle checkpoints (Cdc25A OE), often associated with malignant transformation. We propose that the replication stress observed early after oncogene induction is tolerated in CycE-overexpressing cells by means of controlled processing of replication intermediates during a transient, checkpoint-mediated delay in G2/M (Lukas et al., 2011). This transient arrest limits DNA breakage, as ATR inhibition under these conditions leads to an increase in CycE-induced DSBs (Fig. S3, J–L). However, the accumulation of “DNA lesions” upon prolonged CycE OE (Figs. 1 and S1) may eventually lead to a persistent G2/M arrest, extensive processing of replication intermediates, inaccurate chromosome segregation (e.g., micronuclei; Fig. S3 G), and abortive attempts to restart DNA replication (Fig. 5 A; Di Micco et al., 2006; Davoli et al., 2010; Crasta et al., 2012). In contrast, Cdc25A-overexpressing cells—prototypic of checkpoint-deficient cells experiencing oncogenic stress—display constitutive CDK1 hyperactivation and are therefore unable to restrain mitotic processing, which is initiated together with the earliest manifestations of replication stress. Thus, they rapidly incur extensive DNA breakage, strong DDR activation, and replicative arrest. We propose that the crucial role of the DDR as an anticancer barrier results from its molecular function in the tolerance of oncogene-induced replication stress, by ensuring controlled processing of unusual RI and thus preventing excessive chromosomal breakage.
Finally, our work provides important mechanistic insight in the cellular activities involved in oncogene-induced chromosomal breakage. Combining PFGE and EM analysis, we show that MUS81/EME1—a cell cycle–regulated HJ resolvase (Matos et al., 2011)—is a major contributor to Cdc25A-induced DSBs by processing reversed forks (Fig. 5 B). These data demonstrate that HJs at replication forks are in vivo substrates of this nuclease, despite the weak cleavage activity in vitro (Taylor and McGowan, 2008). Interestingly, at least a fraction of reversed forks displays ss-nicks or gaps at the branch point (Figs. 4 J and S1 A), which are known to enhance susceptibility to cleavage by MUS81 (Schwartz and Heyer, 2011). Furthermore, the observation that MUS81 controls the abundance of reversed forks also in unperturbed conditions suggests that these structures are formed even in the absence of exogenous stress and that their controlled processing is required for genome maintenance in every cell cycle.
Materials and methods
Cell culture, treatments, and transfections
U2OS-derived clones carrying inducible copies of CycE and Cdc25A were grown in DMEM + 10% FCS supplemented with 4 µg/ml tetracycline. Oncogene expression was induced by washing off tetracycline. MRC5 cells were grown in DMEM + 10% FCS. For inhibition of CDK1, RO-3306 (#217699; EMD Millipore) was added 3 h after oncogene induction at a final concentration of 9 µM. Tetracycline was from Sigma-Aldrich (T7660). For inhibition of ATR, the ATR inhibitor ETP-46464 (kindly provided by O. Fernandez-Capetillo, CNIO, Madrid, Spain) was added for 12 h before collection at a final concentration of 2 µM. For oncogene OE in MRC5 cells, cells were transfected with pBabe (empty vector), or plasmid encoding Cdc25A or CycE (kindly provided by J. Lukas, Center for Protein Research, Copenhagen, Denmark), respectively, at the indicated time points before collection using FuGENE6 (Promega) according to the manufacturer’s instructions. For depletion experiments, cells were transfected 72 h before oncogene induction with the indicated siRNA using RNAiMax (Invitrogen) according to the manufacturer’s instructions. siLUC (33 nM; 5′-GGUACGCGGAAUACUUCGAdTdT-3′), siMUS81#1 (33 nM; 5′-CAGCCCUGGUGGAUCGAUAdTdT-3′), and siMUS81#2 (80 nM; 5′-CAGGAGCCAUCAAGAAUAAdTdT-3′).
Flow cytometry
Cell cycle analysis by propidium iodide staining was performed as described previously (Toller et al., 2011). In brief, cells were fixed with ice-cold 70% ethanol, washed with PBS, pH 7.4, and DNA was stained with 25 µg/ml propidium iodide (Sigma-Aldrich). For flow cytometric analysis for γ-H2AX/EdU/DAPI, cells were labeled for 30 min with 10 µM EdU, harvested, and fixed for 10 min with 4% formaldehyde/PBS. Cells were washed with 1% BSA/PBS, pH 7.4, permeabilized with 0.5% saponin/1% BSA/PBS, and stained with anti–γ-H2AX antibody (#05-636; EMD Millipore) for 2 h, followed by incubation with a suitable secondary antibody for 30 min. Incorporated EdU was labeled according to the manufacturer’s instructions (#C35002; Invitrogen). For flow cytometric analysis for γ-H2AX/MPM-2/DAPI, cells were fixed and permeabilized as described above, followed by incubation with antibodies against γ-H2AX (#9718; Cell Signaling Technology) and MPM-2 (#05-368; EMD Millipore) and suitable secondary antibodies. In both assays, DNA was stained with 1 µg/ml DAPI. Samples were measured on a Cyan ADP flow cytometer (Beckman Coulter) and analyzed with Summit software v4.3 (Beckman Coulter).
Pulse-field gel electrophoresis, single-cell microscopy, and antibodies
Pulse-field gel electrophoresis was performed as reported previously (Toller et al., 2011). In brief, cells were embedded in a 0.8% agarose plug (2.5 × 105 cells/plug), digested in lysis buffer (100 mM EDTA, 1% [wt/vol] sodium lauryl sarcosyne, 0.2% [wt/vol] sodium deoxycholate, and 1 mg/ml proteinase K) at 37°C for 48 h, and washed in 10 mM Tris-HCl, pH 8.0, and 100 mM EDTA. Electrophoresis was performed at 14°C in 0.9% (wt/vol) Pulsed Field Certified Agarose (Bio-Rad Laboratories) containing Tris-borate/EDTA buffer in a CHEF DR III apparatus (9 h, 120°, 5.5 V/cm, 30–18 s switch time; 6 h, 117°, 4.5 V/cm, 18–9 s switch time; 6 h, 112°, 4 V/cm, 9–5 s switch time; Bio-Rad Laboratories). The gel was stained with ethidium bromide and imaged on an Alpha Innotech Imager. For single-cell immunostaining, cells were fixed with 4% formaldehyde/PBS, stained for γ-H2AX and 53BP1 as indicated, detected by appropriate secondary antibodies, and mounted with Vectashield (Vector Laboratories). Cells were imaged using a microscope (model DMRB; Leica) equipped with a camera (model DFC360 FX; Leica). Images were taken at 60×, using Leica Application Suite 3.3.0. The following primary antibodies were used: γ-H2AX (#05-636; EMD Millipore), 53BP1 (sc-22760; Santa Cruz Biotechnology, Inc.), CycE (sc-198; Santa Cruz Biotechnology, Inc.), Cdc25A (sc-7389; Santa Cruz Biotechnology, Inc.), CHK1 pS345 (#2348; Cell Signaling Technology), CHK1 (sc-8408; Santa Cruz Biotechnology, Inc.), KAP1-pS824 (A300-767A; Bethyl Laboratories, Inc.), KAP1 (A300-274A; Bethyl Laboratories, Inc.), TFIIH (sc-293; Santa Cruz Biotechnology, Inc.), MUS81 (M1445; Sigma-Aldrich), and β-tubulin (sc-5274; Santa Cruz Biotechnology, Inc.). Secondary antibodies were Alexa Fluor conjugates (Alexa Fluor 488, 594, and 647; Invitrogen).
DNA fiber spreadings
Cells were sequentially pulse-labeled with 30 µM IdU and 250 µM CldU for 20 min each and harvested. Cells were then lysed and DNA fibers stretched onto glass slides, as described previously (Ray Chaudhuri et al., 2012). In brief, the fibers were denatured with 2.5 M HCl for 1 h, washed with PBS, and blocked with 0.2% Tween 20 in 2% BSA/PBS. CldU and IdU tracks were revealed with anti-BrdU antibodies recognizing CldU (ab6326; Abcam) and IdU (347580; BD), respectively, and appropriate secondary antibodies. Images were acquired with a microscope (model IX81; Olympus), CellR software (Olympus), and an Orca camera (Hamamatsu Photonics). Statistical analysis was performed using Prism (GraphPad Software).
Electron microscopic analysis of genomic DNA
In vivo psoralen cross-linking, isolation of total genomic DNA, and enrichment of the replication intermediates and their EM visualization were performed as described previously (Ray Chaudhuri et al., 2012; Neelsen et al., 2013). In brief, cells were harvested, and genomic DNA was cross-linked by two rounds of incubation in 10 µM 4,5′,8-trimethylpsoralen and 2 min of irradiation with 366-nm UV light. Cells were lysed, and genomic DNA was isolated from the nuclei by proteinase K digestion and phenol-chloroform extraction. Purified DNA was digested with PvuII and replication intermediates were enriched on a BND cellulose column. EM samples were prepared by spreading the DNA on carbon-coated grids and visualized by platinum rotary shadowing. Images were acquired on a microscope (G2 Spirit; FEI Tecnai) and analyzed with ImageJ (National Institutes of Health). Statistical analysis was performed using Prism.
Online supplemental material
Fig. S1 shows reversed replication forks and replication intermediates with exposed ssDNA observed upon OE of Cdc25A and CycE, and statistics on the accumulation of ssDNA regions under these conditions. Fig. S2 shows replication fork slow-down, accumulation of unusual replication intermediates, DSB formation, and DDR activation upon OE of Cdc25A and CycE in MRC5 cells. Fig. S3 illustrates the different H2AX phosphorylation patterns detected upon oncogene OE in FACS and single-cell IF, shows IF data on DDR markers (γ-H2AX and 53BP1), and accumulation of micronuclei and cells with ≥4n DNA in oncogene-overexpressing cells, FACS profiles for samples in Fig. 4, and the effect of ATR inhibition on CycE-overexpressing cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212058/DC1.
Supplementary Material
Acknowledgments
We are grateful to E. Petermann for sharing information before publication and to S. Ferrari and all members of the Lopes laboratory for helpful discussions. We thank O. Fernandez-Capetillo and J. Lukas for reagents, and the Center for Microscopy and Image Analysis of the University of Zurich for technical assistance.
This work was supported by Swiss National Science Foundation grants (PP0033-114922, PP00P3-1352), Krebsliga Zurich, and Research Priority Program on Systems Biology of the University of Zurich.
Footnotes
Abbreviations used in this paper:
- DDR
- DNA damage response
- DSB
- DNA double-strand break
- EdU
- 5-ethynyl-2′-deoxyuridine
- HJ
- Holliday junction
- HR
- homologous recombination
- IF
- immunofluorescence
- OE
- overexpression
- PFGE
- pulse-field gel electrophoresis
- RF
- reversed replication fork
- RI
- replication intermediate
- ssDNA
- single-stranded DNA
References
- Bartek J., Bartkova J., Lukas J. 2007. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 26:7773–7779 10.1038/sj.onc.1210881 [DOI] [PubMed] [Google Scholar]
- Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 434:864–870 10.1038/nature03482 [DOI] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Beck H., Nähse V., Larsen M.S., Groth P., Clancy T., Lees M., Jørgensen M., Helleday T., Syljuåsen R.G., Sørensen C.S. 2010. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J. Cell Biol. 188:629–638 10.1083/jcb.200905059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., Carotenuto W., Cocito A., Doksani Y., Klein H., Gómez-González B., et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 146:233–246 10.1016/j.cell.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Lai M.S., Foiani M. 2012. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell. 45:710–718 10.1016/j.molcel.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 145:435–446 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F.M., Tsao T.Y., Fowler S.K., Rao P.N. 1983. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. USA. 80:2926–2930 10.1073/pnas.80.10.2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., Denchi E.L., de Lange T. 2010. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 141:81–93 10.1016/j.cell.2010.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Feraudy S., Revet I., Bezrookove V., Feeney L., Cleaver J.E. 2010. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 107:6870–6875 10.1073/pnas.1002175107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P.G., Bensimon A., et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 444:638–642 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Domínguez-Kelly R., Martín Y., Koundrioukoff S., Tanenbaum M.E., Smits V.A., Medema R.H., Debatisse M., Freire R. 2011. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 194:567–579 10.1083/jcb.201101047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Ying C.Y., Grandori C., Ruggiero L., Chen B., Li M., Galloway D.A., Gu W., Gautier J., Dalla-Favera R. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature. 448:445–451 10.1038/nature05953 [DOI] [PubMed] [Google Scholar]
- Forment J.V., Blasius M., Guerini I., Jackson S.P. 2011. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS ONE. 6:e23517 10.1371/journal.pone.0023517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S., Belotserkovskaya R., Jackson S.P. 2010. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190:197–207 10.1083/jcb.200911156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Jr, Kastrinakis N.G., Levy B., et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 434:907–913 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Modesti M., Maas A., Wyman C., Essers J., Kanaar R. 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 25:4921–4932 10.1038/sj.emboj.7601344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., Kanaar R. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Higgins N.P., Kato K., Strauss B. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417–425 10.1016/0022-2836(76)90156-X [DOI] [PubMed] [Google Scholar]
- Ichijima Y., Yoshioka K., Yoshioka Y., Shinohe K., Fujimori H., Unno J., Takagi M., Goto H., Inagaki M., Mizutani S., Teraoka H. 2010. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS ONE. 5:e8821 10.1371/journal.pone.0008821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Pombo A. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285–1295 10.1083/jcb.140.6.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.M., Mortusewicz O., Afzal I., Lorvellec M., Garcia P., Helleday T., Petermann E. 2012. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 10.1038/onc.2012.387 [DOI] [PubMed] [Google Scholar]
- Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøfte M., Chan K.L., Hickson I.D., Bartek J., Lukas J. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- Mailand N., Falck J., Lukas C., Syljuâsen R.G., Welcker M., Bartek J., Lukas J. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science. 288:1425–1429 10.1126/science.288.5470.1425 [DOI] [PubMed] [Google Scholar]
- Matos J., Blanco M.G., Maslen S., Skehel J.M., West S.C. 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 147:158–172 10.1016/j.cell.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus K.J., Hendzel M.J. 2005. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell. 16:5013–5025 10.1091/mbc.E05-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M., Mercurio C., Dominguez J., Goubin F., Draetta G.F. 2000. Human Cdc25 A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 1:71–79 10.1093/embo-reports/kvd018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I., Nicolai S., Baldari S., Crescenzi M., Bignami M., Franchitto A., Pichierri P. 2013. The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene. 32:610–620 10.1038/onc.2012.80 [DOI] [PubMed] [Google Scholar]
- Murga M., Bunting S., Montaña M.F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P.J., Nussenzweig A., Fernandez-Capetillo O. 2009. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 41:891–898 10.1038/ng.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K.J., Ray Chaudhuri A., Follonier C., Herrador R., Lopes M. 2013. Visualization and interpretation of eukaryotic DNA replication intermediates by electron microscopy in vivo. Methods Mol. Biol. In press [DOI] [PubMed] [Google Scholar]
- Postow L., Ullsperger C., Keller R.W., Bustamante C., Vologodskii A.V., Cozzarelli N.R. 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 276:2790–2796 10.1074/jbc.M006736200 [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19:417–423 10.1038/nsmb.2258 [DOI] [PubMed] [Google Scholar]
- Schwartz E.K., Heyer W.D. 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 120:109–127 10.1007/s00412-010-0304-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., Foiani M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Taylor E.R., McGowan C.H. 2008. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc. Natl. Acad. Sci. USA. 105:3757–3762 10.1073/pnas.0710291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L.I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J.R., Fernandez-Capetillo O. 2011. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18:721–727 10.1038/nsmb.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toller I.M., Neelsen K.J., Steger M., Hartung M.L., Hottiger M.O., Stucki M., Kalali B., Gerhard M., Sartori A.A., Lopes M., Müller A. 2011. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA. 108:14944–14949 10.1073/pnas.1100959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 103:10660–10665 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Chen Y., Dutta A. 2004. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24:7140–7150 10.1128/MCB.24.16.7140-7150.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.