Abstract
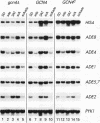
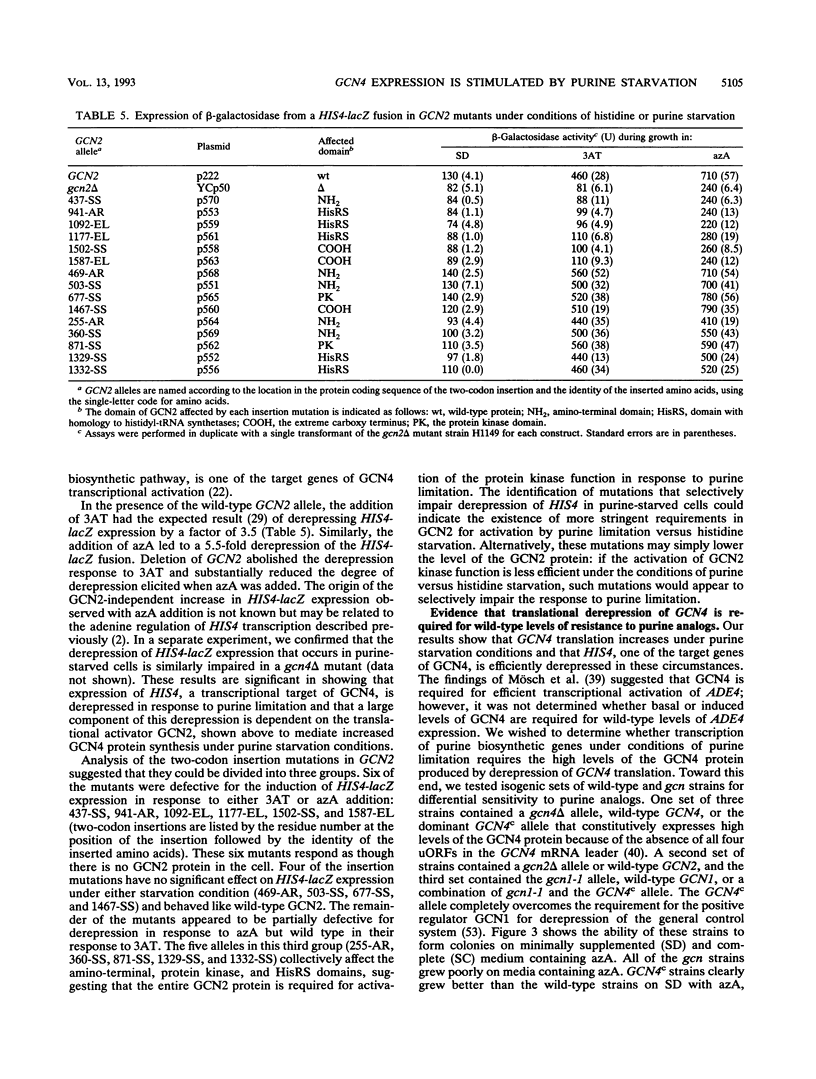
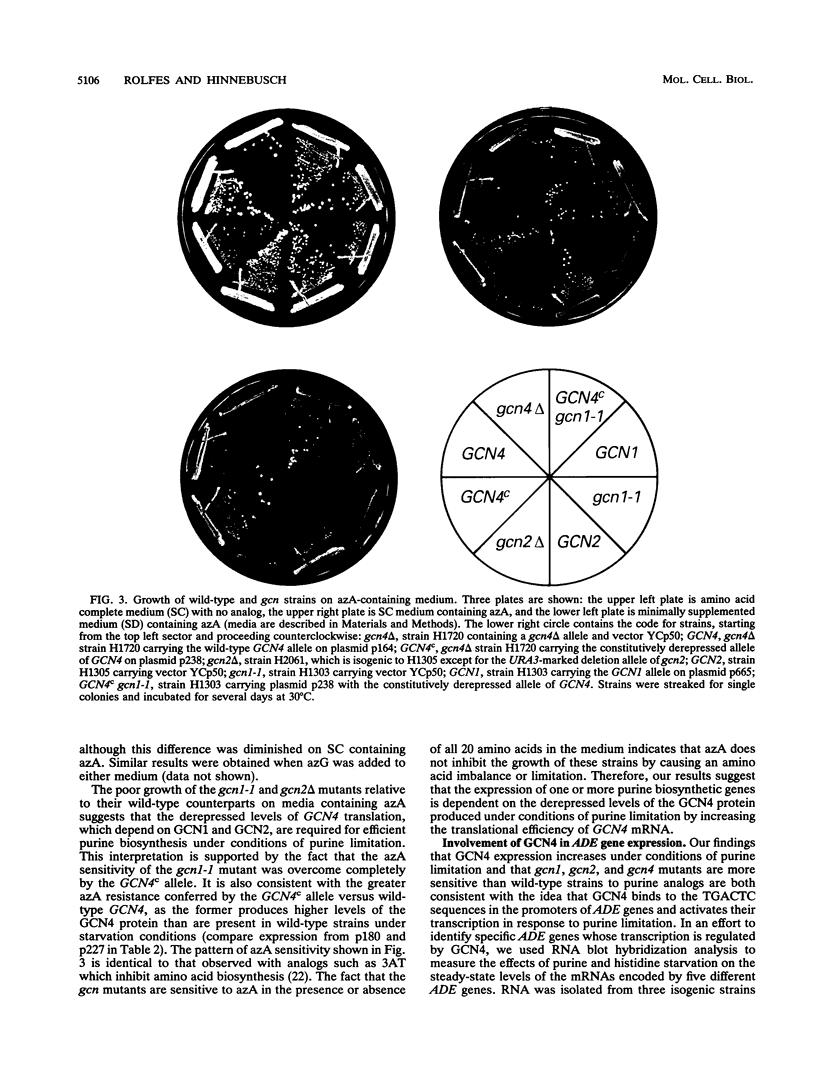
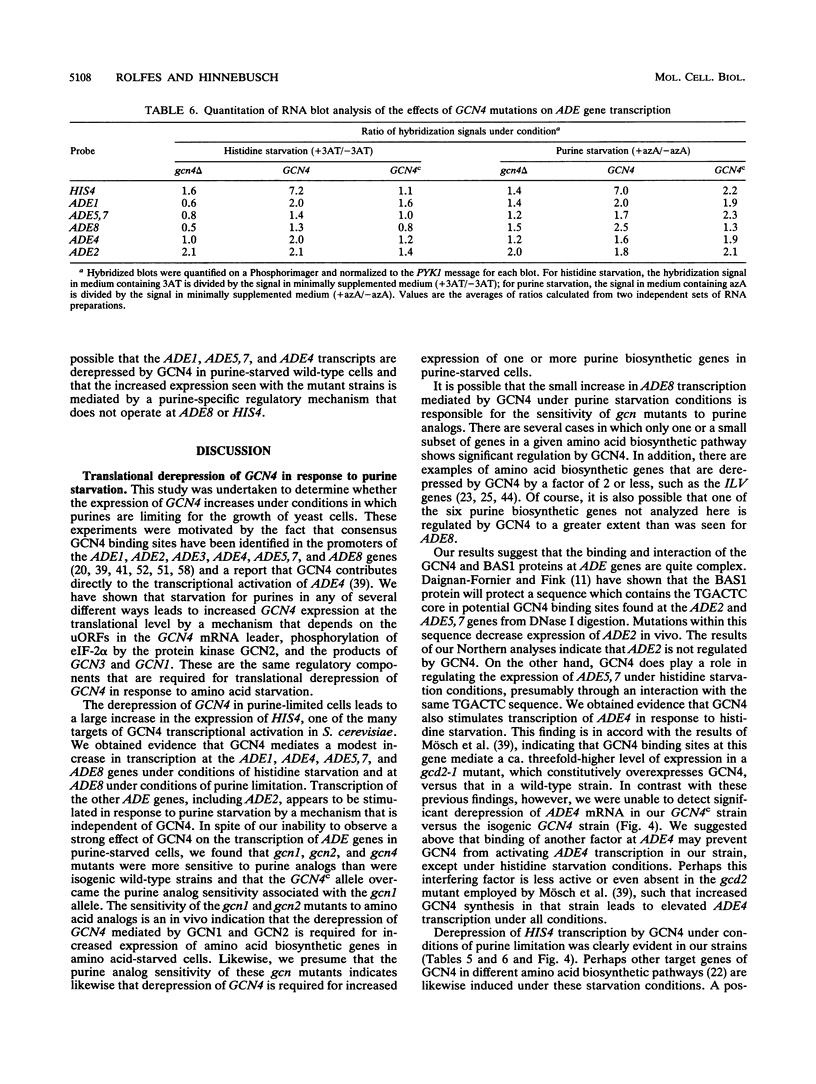
The transcriptional activator protein GCN4 is responsible for increased transcription of more than 30 different amino acid biosynthetic genes in response to starvation for a single amino acid. This induction depends on increased expression of GCN4 at the translational level. We show that starvation for purines also stimulates GCN4 translation by the same mechanism that operates in amino acid-starved cells, being dependent on short upstream open reading frames in the GCN4 mRNA leader, the phosphorylation site in the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2 alpha), the protein kinase GCN2, and translational activators of GCN4 encoded by GCN1 and GCN3. Biochemical experiments show that eIF-2 alpha is phosphorylated in response to purine starvation and that this reaction is completely dependent on GCN2. As expected, derepression of GCN4 in purine-starved cells leads to a substantial increase in HIS4 expression, one of the targets of GCN4 transcriptional activation. gcn mutants that are defective for derepression of amino acid biosynthetic enzymes also exhibit sensitivity to inhibitors of purine biosynthesis, suggesting that derepression of GCN4 is required for maximal expression of one or more purine biosynthetic genes under conditions of purine limitation. Analysis of mRNAs produced from the ADE4, ADE5,7, ADE8, and ADE1 genes indicates that GCN4 stimulates the expression of these genes under conditions of histidine starvation, and it appeared that ADE8 mRNA was also derepressed by GCN4 in purine-starved cells. Our results indicate that the general control response is more global than was previously imagined in terms of the type of nutrient starvation that elicits derepression of GCN4 as well as the range of target genes that depend on GCN4 for transcriptional activation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K. T., Styles C., Fink G. R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987 Aug 21;237(4817):874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Mar;13(3):1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Bushman J. L., Boal T. R., Hinnebusch A. G. A protein complex of translational regulators of GCN4 mRNA is the guanine nucleotide-exchange factor for translation initiation factor 2 in yeast. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5350–5354. doi: 10.1073/pnas.90.11.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Foiani M., Hannig E. M., Hinnebusch A. G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991 Jun;11(6):3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M. J., Galpine A., Austin S. A., Panniers R., Henshaw E. C., Duncan R., Hershey J. W., Pollard J. W. Regulation of polypeptide chain initiation in Chinese hamster ovary cells with a temperature-sensitive leucyl-tRNA synthetase. Changes in phosphorylation of initiation factor eIF-2 and in the activity of the guanine nucleotide exchange factor GEF. J Biol Chem. 1987 Jan 15;262(2):767–771. [PubMed] [Google Scholar]
- Cooley L., Appel B., Söll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5' terminus of histidine tRNA. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B., Fink G. R. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C. J., Hutchison C. A., 3rd A directed DNA sequencing strategy based upon Tn3 transposon mutagenesis: application to the ADE1 locus on Saccharomyces cerevisiae chromosome I. Nucleic Acids Res. 1991 Oct 25;19(20):5731–5738. doi: 10.1093/nar/19.20.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Chen J. J., Barber G. N., Cigan A. M., Feng L., Donahue T. F., London I. M., Katze M. G., Hinnebusch A. G. Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992 Feb 7;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., Pabich E. K., Valavicius B. C. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988 Aug 26;54(5):621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. The Saccharomyces cerevisiae ADE5,7 protein is homologous to overlapping Drosophila melanogaster Gart polypeptides. J Mol Biol. 1986 Aug 20;190(4):519–528. doi: 10.1016/0022-2836(86)90238-x. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Transcriptional and translational regulation of gene expression in the general control of amino-acid biosynthesis in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1990;38:195–240. doi: 10.1016/s0079-6603(08)60712-6. [DOI] [PubMed] [Google Scholar]
- Holmberg S., Petersen J. G. Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1988 Mar;13(3):207–217. doi: 10.1007/BF00387766. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Safer B. Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J Biol Chem. 1983 Mar 10;258(5):3402–3408. [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCOLLISTER R. J., GILBERT W. R., Jr, ASHTON D. M., WYNGAARDEN J. B. PSEUDOFEEDBACK INHIBITION OF PURINE SYNTHESIS BY 6-MERCAPTOPURINE RIBONUCLEOTIDE AND OTHER PURINE ANALOGUES. J Biol Chem. 1964 May;239:1560–1563. [PubMed] [Google Scholar]
- Marton M. J., Crouch D., Hinnebusch A. G. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol. 1993 Jun;13(6):3541–3556. doi: 10.1128/mcb.13.6.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for beta-galactosidase activity. Anal Biochem. 1989 Aug 15;181(1):40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Delforge J. Role of transfer ribonucleic acids in the regulation of several biosyntheses in Saccharomyces cerevisiae. Eur J Biochem. 1976 Aug 16;67(2):335–339. doi: 10.1111/j.1432-1033.1976.tb10696.x. [DOI] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2767–2773. doi: 10.1128/mcb.4.12.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C. M., Hinnebusch A. G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Myasnikov A. N., Sasnauskas K. V., Janulaitis A. A., Smirnov M. N. The Saccharomyces cerevisiae ADE1 gene: structure, overexpression and possible regulation by general amino acid control. Gene. 1991 Dec 20;109(1):143–147. doi: 10.1016/0378-1119(91)90600-g. [DOI] [PubMed] [Google Scholar]
- Mäntsälä P., Zalkin H. Glutamine nucleotide sequence of Saccharomyces cerevisiae ADE4 encoding phosphoribosylpyrophosphate amidotransferase. J Biol Chem. 1984 Jul 10;259(13):8478–8484. [PubMed] [Google Scholar]
- Mösch H. U., Scheier B., Lahti R., Mäntsäla P., Braus G. H. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J Biol Chem. 1991 Oct 25;266(30):20453–20456. [PubMed] [Google Scholar]
- Nagy M. Regulation of the biosynthesis of purine nucleotides in Schizosaccharomyces pombe. I. Properties of the phosphoribosylpyrophosphate: glutamine amidotransferase of the wild strain and of a mutant desensitized towards feedback modifiers. Biochim Biophys Acta. 1970 Mar 18;198(3):471–481. doi: 10.1016/0005-2744(70)90125-7. [DOI] [PubMed] [Google Scholar]
- Petersen J. G., Holmberg S. The ILV5 gene of Saccharomyces cerevisiae is highly expressed. Nucleic Acids Res. 1986 Dec 22;14(24):9631–9651. doi: 10.1093/nar/14.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek R. C., Hinnebusch A. G. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek R. C., Vazquez de Aldana C. R., Jackson B. M., Freeman B., Hinnebusch A. G. Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol. 1992 Dec;12(12):5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimans M., Goumans H., Amesz H., Benne R., Voorma H. O. Regulation of protein synthesis in eukaryotes. Mode of action of eRF, an eIF-2-recycling factor from rabbit reticulocytes involved in GDP/GTP exchange. Eur J Biochem. 1984 Nov 15;145(1):91–98. doi: 10.1111/j.1432-1033.1984.tb08526.x. [DOI] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staben C., Rabinowitz J. C. Nucleotide sequence of the Saccharomyces cerevisiae ADE3 gene encoding C1-tetrahydrofolate synthase. J Biol Chem. 1986 Apr 5;261(10):4629–4637. [PubMed] [Google Scholar]
- Stotz A., Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990 Oct 30;95(1):91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. S., Matts R. L., Petryshyn R., London I. M. Distribution of reversing factor in reticulocyte lysates during active protein synthesis and on inhibition by heme deprivation or double-stranded RNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6998–7002. doi: 10.1073/pnas.81.22.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989 Jun 16;57(6):947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Jackson B. M., Hinnebusch A. G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Ramirez M., Jackson B. M., Hinnebusch A. G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990 Jun;10(6):2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., DiMartino J. F., Anderson R. W., Lusnak K., Hilbert D., Fogel S. A DNA sequence conferring high postmeiotic segregation frequency to heterozygous deletions in Saccharomyces cerevisiae is related to sequences associated with eucaryotic recombination hotspots. Mol Cell Biol. 1988 Mar;8(3):1253–1258. doi: 10.1128/mcb.8.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7515–7519. doi: 10.1073/pnas.86.19.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]