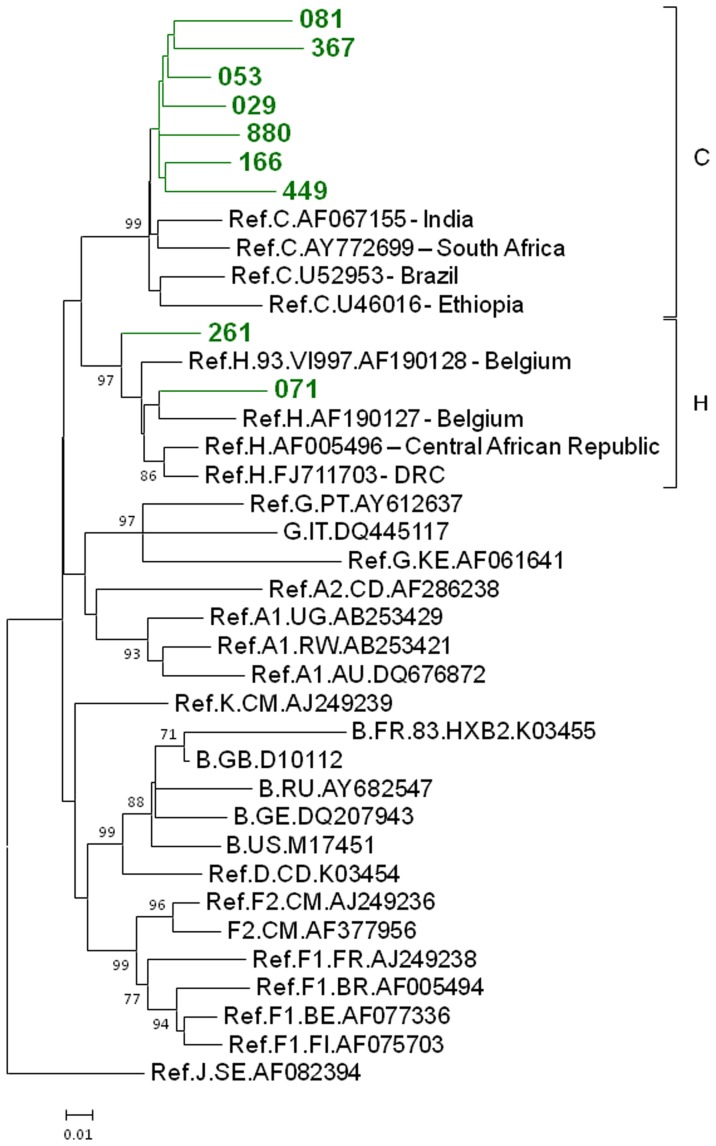
Figure 3. Inferred phylogenetic relationships of Bulgarian HIV-1 subtypes C and H.
Tree structure was inferred using maximum likelihood analysis of polymerase sequences implemented in MEGA5. Support for each node was determined using 1,000 bootstrap replications with only values ≥70 shown. Scale bar indicates the number of nucleotide substitutions per site. Antiretroviral resistance-associated mutations were stripped from the alignments. Nearly identical tree topologies were also obtained with Bayesian analysis. The 690-bp alignment consisted of 9 HIV-1 C and H strains from Bulgaria and 29 Group M reference sequences from the Los Alamos HIV database. The tree was rooted by using an HIV-1 subtype J strain as the outgroup. Bulgarian sequences are shown using green branches and taxon names.