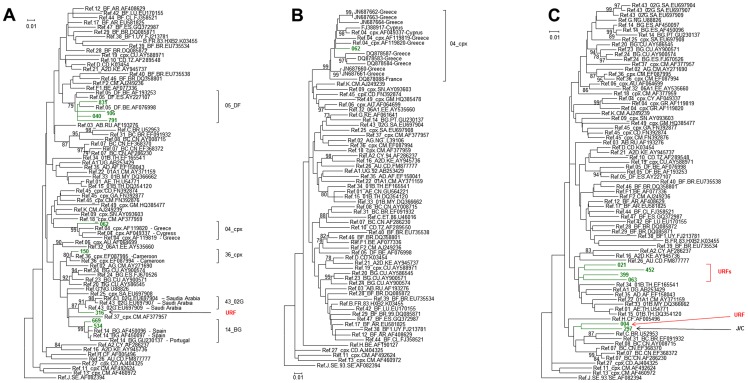
Figure 6. Inferred phylogenetic relationships of Bulgarian HIV-1 subtypes.
Tree structure was inferred using maximum likelihood analysis of polymerase sequences implemented in MEGA5. Support for each node was determined using 1,000 bootstrap replications with only values ≥70 shown. Scale bar indicates the number of nucleotide substitutions per site. Antiretroviral resistance-associated mutations were stripped from the alignments. Nearly identical tree topologies were also obtained with Bayesian analysis. (A) Multiple Circulating and unique recombinant forms (CRF and URF, respectively). The 696-bp alignment was composed of one CRF04_cpx, four CRF05_DF, two CRF14_BG, and one CRF36_cpx strains and one CRF43_02G-like strain from Bulgaria and 71 related reference sequences from the Los Alamos HIV database. The tree was rooted by using an HIV-1 subtype J strain as the outgroup. Bulgarian sequences are shown using green branches and taxon names. (B) CRF04_cpx. The 1043-bp alignment was composed of one HIV-1 CRF04_cpx1 strain from Bulgaria and 69 reference sequences from the Los Alamos HIV database and related sequences identified by BLAST. The tree was rooted by using an HIV-1 subtype J sequence as the outgroup. Bulgarian sequence is depicted using a green branch and taxon name. (C) Identification of novel HIV-1 URFs in Bulgaria. The 696-bp alignment was composed of six HIV-1 URF strains from Bulgaria and 68 reference sequences from the Los Alamos HIV database. The tree was rooted by using an HIV-1 subtype J strain as the outgroup. Bulgarian sequences are shown using green branches and taxon names. Bulgarian sequence #297 was shown by SimPlot analysis to be a novel recombinant of subtype J and C sequences.