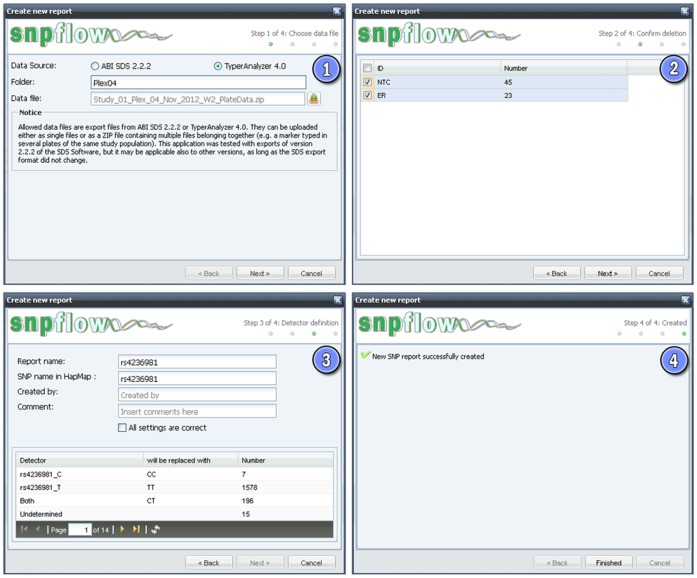
Figure 3. The SNPflow data analysis wizard.
This figure shows the 4 steps of the SNPflow data analysis wizard. In brief, the first step allows the uploading of either SDS files (ABI 7900HT) or TyperAnalyzer 4.0 (MassARRAY) files. For TyperAnalyzer files, a second input field asks for the name of the result group under which results shall be grouped. The second step checks the validity of the IDs found in the uploaded and reports IDs, which are not found in the study definition and shall thus be deleted. Step 3 converts the found genotype designations to standard genotypes and allows entering the name of the result, the operator name, an optional comment and the rs-number of the SNP in HapMap and the 1000 Genomes Project. This is useful in case that a SNP ID changed between the current dbSNP release and the dbSNP release used in HapMap, as happened for some SNPs. For convenience, the result name is automatically pre-set as HapMap lookup. In multiplex assays, pagination allows navigating through the single SNPs. For quality reasons a manual confirmation of each conversion is required (Checkbox “All settings are correct”). Finally, step 4 confirms the successful analysis and brings the user to the report. Exemplary abbreviation used in this figure: NTC, Non-template control; ER, annulled samples (“error”), i.e. DNA samples present on the plates, but known to be flawed. The IDs of these samples were thus replaced with “ER” in order to avoid data collection for these samples.