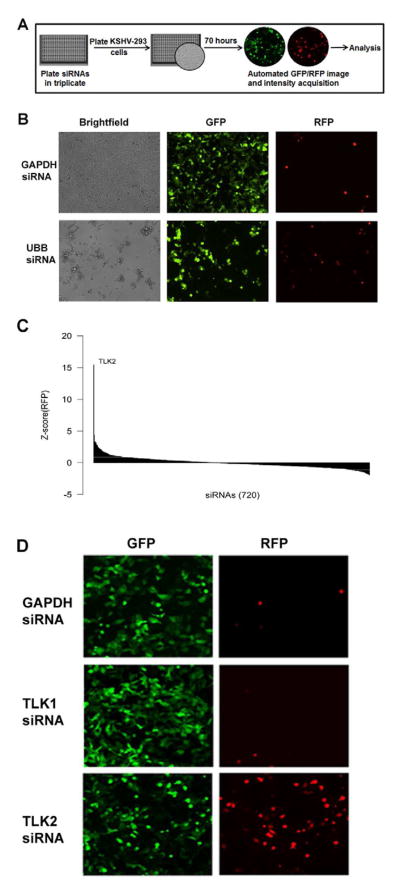
Figure 1. Design of siRNA Screen and Analysis of Data.
A) Schematic of cellular kinome siRNA screen. siRNAs from the Dharmacon SMARTpool kinase siRNA library were loaded into 384-well plates in triplicate. KSHV-293 cells were added to each well containing siRNA and incubated for 70 hours at 37°C. Both RFP and GFP images were then taken for 5 fields per well. B) Control siRNAs demonstrate efficient siRNA transfection in our screen. GAPDH and Ubiquitin B (UBB) siRNAs were reverse transfected at 25nM each into 2500 KSHV-293 cells/well in a 384-well plate. At 70 hours post-transfection brightfield, GFP, and RFP pictures were taken on a fluorescent microscope. C) Statistical analysis of primary hits. A Waterfall plot was used to analyze the data acquired from the screen. It shows the Z-score of the median RFP intensity of each siRNA. D) Reactivation by TLK2 knockdown in KSHV-293 cells. GFP and RFP images of a representative field taken during the screen are shown for the wells containing siRNA targeting GAPDH, TLK1, and TLK2. See also Fig. S1.