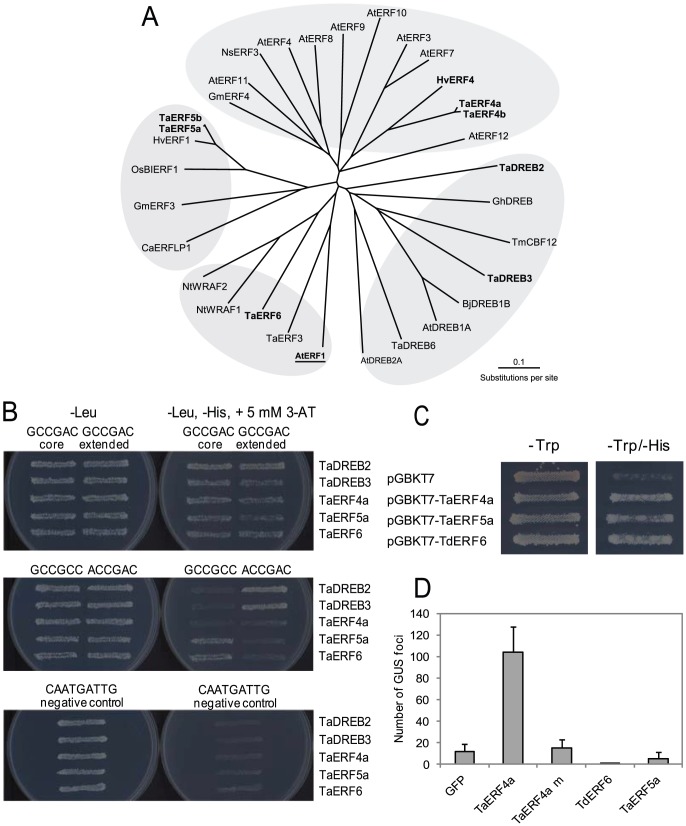
Figure 2. Wheat TFs isolated in Y1H screens and their properties.
(A) An unrooted radial phylogenetic tree of AP2-domain containing TFs from monocotyledonous and dicotyledonous plant species. Amino acid sequences of 32 proteins were aligned with ProMals3D (44) and branch lengths were drawn to scale. Grey shading indicates distinct branches of ERF and DREB TFs. Two-letter prefixes in the sequence identifiers indicate species of origin (Ta = Triticum aestivum; Hv = Hordeum vulgare; Os = Oriza sativa; Gm = Glycine max; At = Arabidopsis thaliana; Bj = Brassica juncea; Gh = Gossypium hirsutum; Nt = Nicotiana tabacum; Ns = Nicotiana sylvestris; Ca = Capsicum annuum). Protein accession numbers are specified in the Materials and Methods. TFs isolated in this work are shown in bold. The Arabidopsis AtERF1 TF was used for construction of 3D models of the AP2 domains of TaERF4a, TaERF5a and TaDREB3, and is shown in bold and underlined. (B) Specificity of recognition of known stress-responsive cis-elements by ERF and DREB TFs detected via a Y1H assay. Growth of yeast on selective medium (-Leu, -His,+5 mM 3-AT) indicates protein-DNA interaction. The cis-element CAATGATTG of the HD-Zip class II TF was used as a negative control. (C) Demonstration of activator properties using ERFs in a Y1H assay. The presence of their own activation domains in the representatives from each subfamily of ERFs supports the activation of the yeast genes and consequent growth of yeast on the selective (-Leu, -Trp, -His, -Ade) medium. (D) Regulation of TdCor410b promoter activity by representatives of each isolated subfamily of ERFs. TFs were tested in a transient expression assay in a wheat cell culture. The pTdCor410b-GUS construct was co-bombarded with pUbi-GFP (GFP; negative control), pUbi-TaERF4a (TaERF4a), pUbi-TaERF4a mutated in the ERF-associated amphiphilic repression (EAR) motif (TaERF4a m), pUbi-TaERF6 (TaERF6), and pUbi-TaERF5a (TaERF5a), and GUS expression in the cultures was quantified (n = 4±SD (P<0.05)).