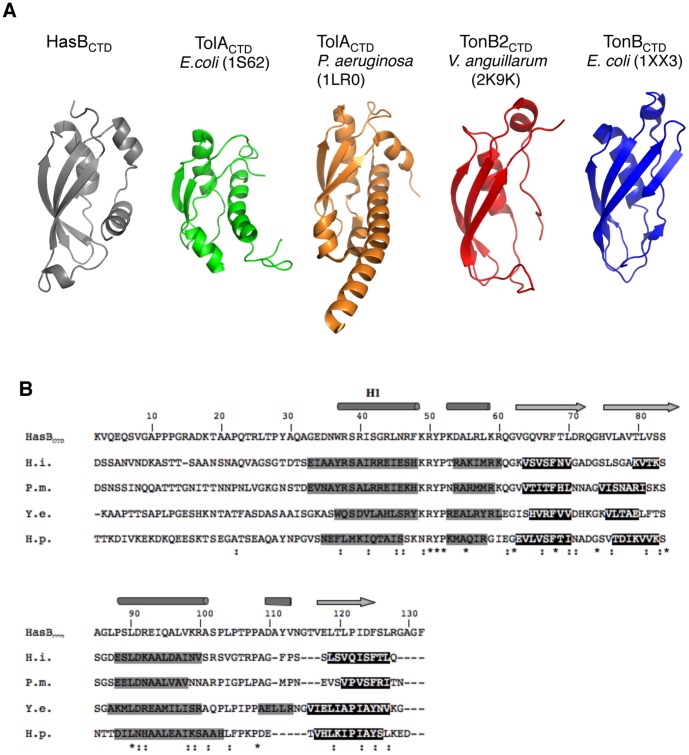
Figure 8. Structural comparison of HasBCTD with TolACTD and TonBCTD. A:
Ribbon representation of available solution structure of TonB and TolA proteins family. B: ClustalW alignments of HasBCTD and TonBCTD with their corresponding secondary structure. Cylinders represent the helices and arrows the β-strands of HasBCTD structure. The secondary structure elements of other proteins are from a consensus prediction generated by NPS@ (Combet C, Blanchet C, Geourjon C, & Deléage, G (2000). NPS@: network protein sequence analysis. Trends. Biochem. Sci. 25, 147–150). Predicted helices and β-strands are respectively presented in black on gray background and in white on black background. (*) conserved residues; (:) similar residues. The sequence numbering is that of HasBCTD. H.i.: residues 143–264 of Haemophilus influenzae TonB (#68248858); P.m.: residues 133–256 of Pasteurella multocida TonB (#33318341); Y.e.: residues 133–256 of Yersinia enterocolitica TonB (#A1JIK8); H.p.: residues 155–279 of Helicobacter pylori TonB1341 (#889856).