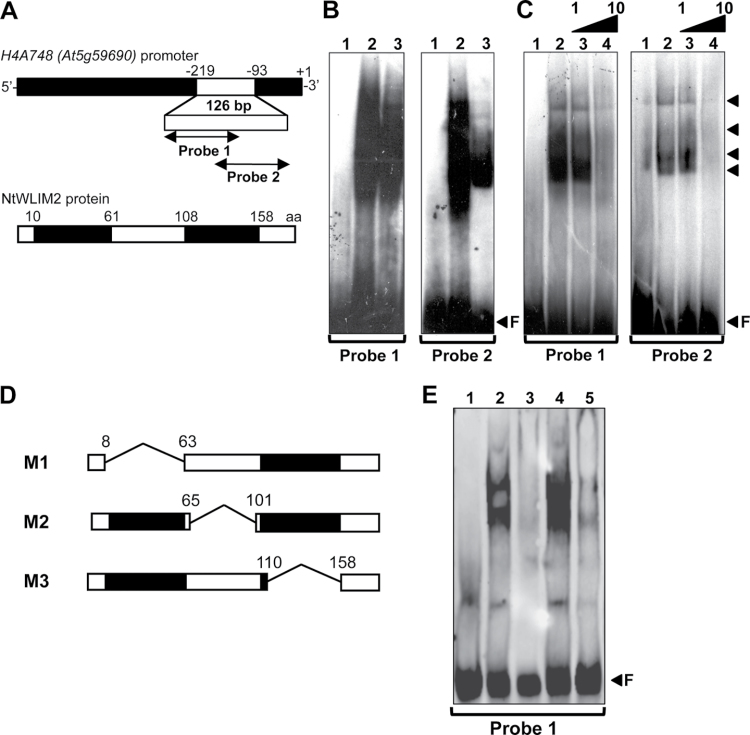
Figure 1.
In Vitro DNA-Binding Ability of NtWLIM2 in Electrophoretic Mobility Shift Assays (EMSAs).(A) Scheme of the Arabidopsis histone H4A748 gene promoter and the derived double-stranded probes. Nucleotide positions are given relative to the transcription start (+1). To avoid disruption of any promoter cis-element, Probe 1 (–219 to –147) and Probe 2 (–161 to –93) were chosen to overlap by 17bp. Schematic representation of the recombinant NtWLIM2 wild-type protein used in the following EMSAs (B, C).(B) EMSA to test the DNA-binding activity of NtWLIM2. One ng of radiolabeled Probe 1 (left panel) or Probe 2 (right panel) was incubated with either DNA-binding buffer (lane 1), nuclear BY-2 protein extract (1μg, lane 2), or purified recombinant NtWLIM2 (3μg, lane 3).(C) Competitive EMSA to check the DNA-binding affinity of NtWLIM2. One ng of radiolabeled Probe 1 (left panel) or Probe 2 (right panel) was incubated with either DNA-binding buffer (lane 1) or purified recombinant NtWLIM2 protein (3μg, lanes 2–4). Lanes 3 and 4 correspond to binding reactions supplemented with equimolar and a 10-fold molar excess of probe, respectively.(D) In addition to the wild-type NtWLIM2, deletion mutants lacking either the LIM1 domain (M1), the interLIM region (M2), or the LIM2 domain (M3) were expressed as 6xHis fusions, affinity-purified, and tested in EMSAs. (E) EMSA with DIG-labeled probes to determine the DNA-binding domains of NtWLIM2. One ng of labeled Probe 1 was incubated with either DNA-binding buffer (lane 1) or 3μg of different recombinant proteins (lanes 2–5): wild-type NtWLIM2 (lane 2) or one of the deletion mutants M1, M2, and M3 (lane 3, 4, and 5, respectively). Arrows mark the migrated free probe (F) and the DNA–protein complexes formed.