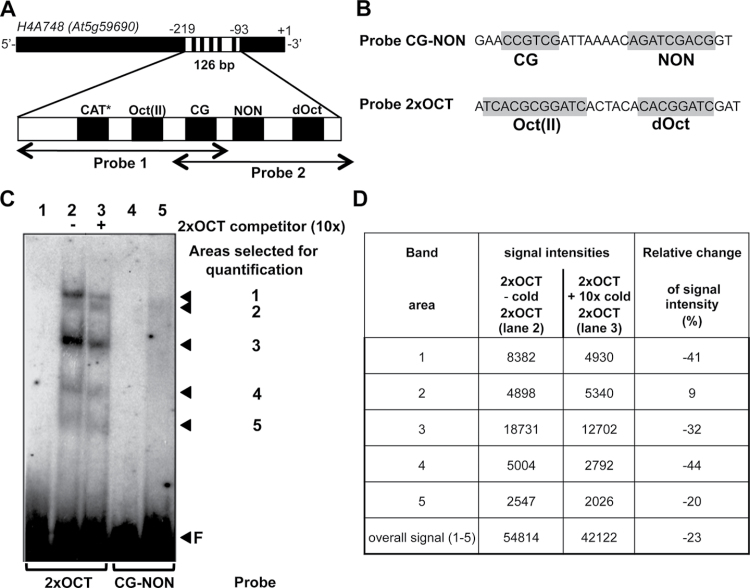
Figure 6.
NtWLIM2 Interaction with Conserved, Plant Histone Promoter cis-Elements. (A) Position of conserved plant histone promoter cis-elements within Arabidopsis H4A748 Probes 1 and 2. Cis-elements located on the reverse strand are marked by an asterisk. CAT, GCAAT-like element; Oct(II), type-II octamer-containing composite element TCACGCGGATC; CG, CCGTC motif; dOct, type II-like, degenerated octamer CACGGATC; NON, nonameric motif AGATCGACG. (B) Conserved plant histone promoter motifs present in Probe 2xOCT and Probe CG–NON (highlighted in gray). (C) EMSA reveals complexes formed between the recombinant NtWLIM2 protein and Arabidopsis pH4A748 promoter cis-elements. One ng of either radiolabeled Probe 2xOCT (lanes 1–3) or CG–NON (lanes 4–5) was incubated with protein storage buffer (lanes 1 and 4, respectively) or 6xHis-tagged affinity-purified NtWLIM2 protein (3μg, lanes 2, 3, and 5). A 10-fold molar excess (lane 3) of cold, double-stranded oligonucleotide 2xOCT was added as a specific competitor to the binding reactions. Arrows mark the migrated free probe (F) and the DNA–protein complexes, whose intensities were quantified by Image J (see (D)). (D) Intensity of shifted bands as determined by ImageJ. Signal intensities for every band are given in the presence and absence of cold 2xOCT probe (band area 1–5). In addition, the sum of the signal over all bands is indicated (overall signal (1–5)). To express the changes in signal intensity produced by the addition of 2xOCT competitor, the ratio of the signal intensity values is calculated for every band and expressed in percent.