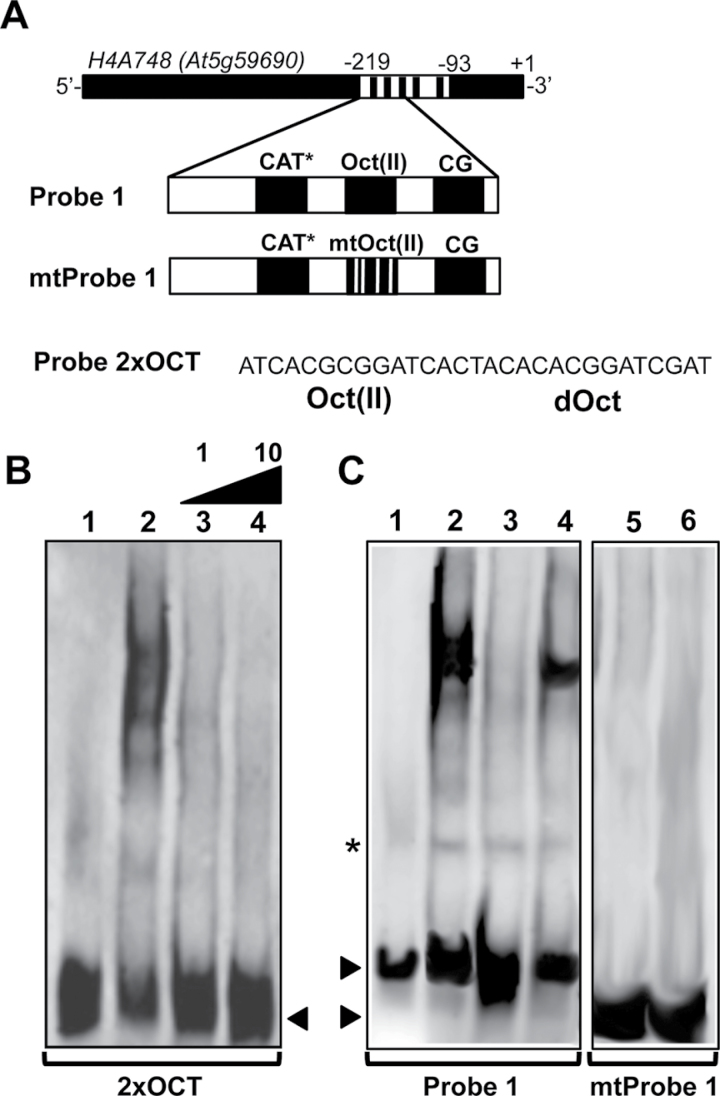
Figure 7.
Determination of NtWLIM2 Binding Specificity by Competitive EMSAs. (A) Scheme of the Arabidopsis histone H4A748 gene promoter and the derived double-stranded Probe 1, mtProbe 1, and 2xOCT used as labeled probes and competitors in the following EMSAs. The mtProbe 1 comprises base exchanges within the conserved Oct(II) element (5’-TCACGCGGATC-3’ changed to 5’-TCACGaaGcTt-3’), which are known to reduce plant histone H3 and H4 promoter activities (Terada et al., 1995; Chaubet et al., 1996). (B) Competition EMSA carried out with 1ng of DIG-labeled 2xOCT probe, which was incubated with either protein storage buffer (lane 1) or purified recombinant NtWLIM2 protein (3μg, lanes 2–4). The shift of the labeled oligo was competed out by the addition of either an equimolar amount or a 10-fold molar excess of unlabeled Probe 1 (lanes 3 and 4, respectively). Arrows mark the migrated free probe. (C) EMSA to analyze the binding specificity of NtWLIM2 to the H4A748 Oct(II) element. A competitive assay (left panel) was performed with 1ng of DIG-labeled Probe 1 (lanes 1–4), which was incubated with either protein storage buffer (lane 1) or recombinant NtWLIM2 protein (3μg, lanes 2–4). A 10-fold molar excess of either cold Probe 1 (lane 3) or cold mtProbe 1 (lane 4) was added as a specific competitor to the binding reactions. In a binding assay (right panel), 1ng of DIG-labeled mtProbe1 (lanes 5–6) was incubated with either protein storage buffer (lane 5) or with 3μg of recombinant NtWLIM2 protein (lane 6). Arrows mark the migrated free probes. An unspecific band shift is indicated by an asterisk.