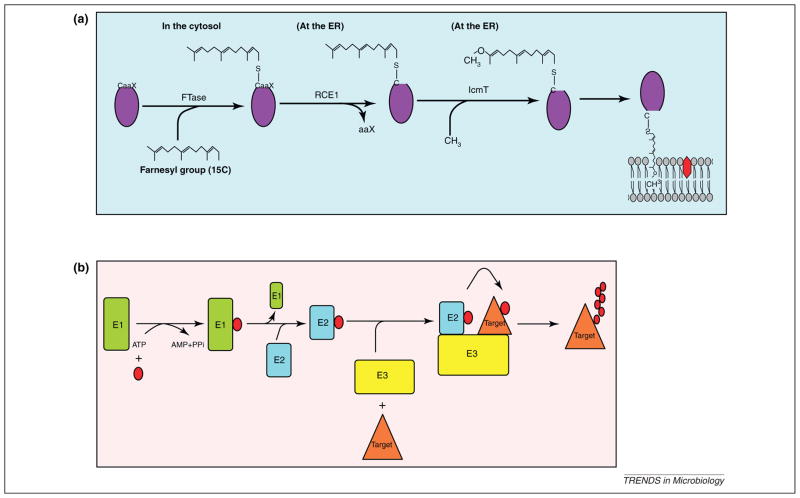
Figure 1.
Evolutionarily conserved eukaryotic pathways exploited by Legionella in amoeba and mammals. (a) Farnesylation modification of eukaryotic proteins. Proteins that contain CaaX motif are recognized by the cytosolic farnesyl transferase (FTase), which modifies the protein by the addition of a 15-carbon farnesyl group to the conserved cysteine residue within the C-terminal CaaX motif. The farnesylated proteins are trafficked to the endoplasmic reticulum (ER) where the aaX tripeptide is cleaved by the Ras-converting enzyme-1 (RCE1) protease, followed by methylation of the farnesyl group by the isoprenyl cysteine carboxyl methyl transferase (IcmT) enzyme. The modified protein is subsequently targeted to specific membranes. (b) The eukaryotic ubiquitination pathway. Ubiquitin modification is an ATP-dependent process involving the sequential action of three enzymes (E1–E3). Ubiquitin activating enzyme (E1) activates ubiquitin (red circle) via the generation of a high energy thioester bond between ubiquitin and an E1 cysteine residue. The activated ubiquitin is then transferred to ubiquitin conjugating enzyme (E2). The final step of the ubiquitination is where substrate-specific ligase (E3) binds simultaneously to E2 and the substrate, resulting in transfer of ubiquitin monomer from the E2 enzyme to a target protein. The fate of the modified protein is determined by the lysine linkages utilized in polyubiquitination.