Abstract
Observing drug responses in the tumor microenvironment in vivo can be technically challenging. As a result, cellular responses to molecularly targeted cancer drugs are often studied in cell culture, which does not accurately represent the behavior of cancer cells growing in vivo. Using high resolution microscopy and fluorescently labeled genetic reporters for apoptosis, we developed an approach to visualize drug-induced cell death at single cell resolution in vivo. Stable expression of the mitochondrial intermembrane protein IMS-RP was established in human breast and pancreatic cancer cells. Automated image analysis was then used to quantify release of IMS-RP into the cytoplasm upon apoptosis and irreversible mitochondrial permeabilization. Both breast and pancreatic cancer cells showed higher basal apoptotic rates in vivo than in culture. To study drug-induced apoptosis, we exposed tumor cells to navitoclax (ABT-263), an inhibitor of Bcl-2, Bcl-xL, and Bcl-w, both in vitro and in vivo. Although the tumors responded to Bcl-2 inhibition in vivo, inducing apoptosis in around 20% of cancer cells, the observed response was much higher in cell culture. Together, our findings demonstrate an imaging technique that can be used to directly visualize cell death within the tumor microenvironment in response to drug treatment.
Keywords: apoptosis, chemotherapy, imaging, breast cancer, pancreatic cancer
Introduction
A major obstacle in the development and monitoring of cancer treatments is determining how multiple biological factors interact to control tumor responses to therapeutic drugs. In vivo imaging represents a powerful, non-invasive tool for revealing cellular responses through time and space. A prevailing challenge, however, has been understanding and quantitating the responses of both individual cancer cells as well as whole tumors to therapeutic drugs; namely, determining the distribution of drugs to cells, the extent and timing of cell death, and the impact of these cell-level responses on tumor regression. Recent advances in intravital microscopy have now enabled high resolution in vivo imaging at the single cell level. This technology potentially not only allows measurement of drug distribution and uptake into tumor cells (pharmacokinetics, PK)[1], but also permits evaluation of downstream responses over different time-scales (pharmacodynamics, PD). To visualize and measure in vitro drug responses in cells, a variety of fluorescent protein (FP) reporters have been developed; these include: caspase cleavable FPs (which can function either as fluorescence resonance energy transfer (FRET)[2] or turn-on probes[3]), mitochondrial outer membrane permeabilization (MOMP) probes[2], senescence markers[4], and autophagy probes[5]. The majority of these reporters, however, have only been tested in model cell lines and thus often fail to adequately reflect the primary tumors on which specific drugs are being tested in trials. Moreover, upon conversion or cell death, many of these reporters exhibit only subtle changes in fluorescence, which tend to be more difficult to measure in vivo due to motion artefacts, and (often) lower signal-to-noise ratios. In contrast, injectable imaging probes (Annexin V, synaptotagmin) have been used primarily for macroscopic imaging without clear microscopic correlates[6-8]. There is thus a clear need for robust reporters that can consistently and reliably detect cell death in common tumor models undergoing treatment with novel targeted drugs.
To address this need, we reasoned that structural analysis of apoptotic tumor cells should be feasible in live animals provided that the resolution is sufficiently high to structurally resolve mitochondria in the cells of live mice. Using a previously described construct[2, 9] and cloning it into a lentiviral vector for greater efficiency, we created breast (MDA-MB-231) and pancreatic (PANC-1) cancer cell lines that stably express the intermembrane space reporter protein (IMS-RP GFP or mCherry). In steady-state cells, mitochondria are seen to contain IMS-RP. Upon apoptosis and irreversible mitochondrial permeabilization, IMS-RP is rapidly released from the spatially conserved intermembrane mitochondrial space into the cytoplasm, where it can be detected and quantified using automated image analysis or visual inspection. Using these cancer models, we not only demonstrate that apoptosis can indeed be visualized in the tumors of live animals, but that apoptotic rates are markedly different in vitro versus in vivo (both temporally and fractionally). We also show that navitoclax monotherapy induces apoptosis in vivo in both breast and pancreatic cancer.
Materials and Methods
Cell culture
MDA-MB-231 human mammary carcinoma cells (ATCC) were grown in RPMI media supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin/streptomycin (Mediatech). PANC-1 human pancreatic adenocarcinoma (ATCC) cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS, 1% L-glutamine, 1% penicillin/streptomycin, and 2% sodium bicarbonate (Lonza). Cells expressing fluorescent reporters were grown in selection medium containing either 3 μg/mL puromycin or 10 μg/mL blasticidin S (Invitrogen). All cells were grown at 37°C and 5% CO2.
Cloning and lentivirus production
Mitochondrial intermembrane space reporter protein (IMS-RP) is an established reporter of mitochondrial outer membrane permeabilization (MOMP); it contains the first 59 amino acids of Smac/DIABLO fused to a fluorescent protein[2]. MOMP is a commitment step during apoptosis, and IMS-RP enables the visualization of this process at single-cell resolution in vitro. pBABE IMS-RP and pBOS H2B-GFP vectors were generously provided by Drs. Peter Sorger and Timothy Mitchison, respectively. IMS-RP and histone H2B were subcloned into the pLVX-AcGFP-N1 or pLVX-mCherry-N1 vectors using In-Fusion cloning (Clontech). Plasmids were verified by DNA sequencing (Genewiz).
For lentivirus production, LentiX-293T cells (Clontech) transfected with pLVX-IMS-RP or pLVX-H2B and Lenti-X HTX Packaging Mix containing lentiviral packaging genes, using the Xfect transfection reagent and following the protocol recommended by the manufacturer (Clontech). Approximately 56-72 hours after transfection, the supernatant was collected, filtered using a 0.45 μm cellulose acetate syringe filter, aliquoted, and then snap frozen in liquid nitrogen for storage at -80°C. All viral supernatants were used immediately upon thawing.
Generation of cell lines expressing reporters
Cells were transduced in 12-well plates using several dilutions of viral supernatant and 4 μg/mL polybrene per well (Invitrogen). Twenty four to forty eight hours following infection, cells were trypsinized and placed into medium containing 3 μg/mL puromycin. When generating double reporter cell lines, double-positive cells were sorted using the FACS Aria II (Becton Dickinson).
In vitro time-lapse microscopy
To determine the kinetics of apoptosis following drug treatment of cell lines in vitro, we used time-lapse microscopy; this enabled us to visualize apoptosis of single cells, within a population of cells. In vitro time-lapse microscopy was carried out using a DeltaVision imaging system (Applied Precision), which consists of an environmental chamber heated to 37°C, with CO2 bubbled through a water bath, an automated Olympus IX70 inverted microscope and a CoolSnap HQ2 CCD camera. Cells were grown and imaged in ibidi 96-well plates (ibidi). Fields of view were chosen based on the equivalence of fluorescence intensity across the cell population (to avoid under/over-exposure of cells) and on adequate cell density. Once fields of view were chosen, cells were treated with a) 25 or 100 μM ABT-263 (Selleck Chemicals) in 0.5 or 2% DMSO, or b) vehicle only, and time-lapse imaging was initiated. Post-acquisition images were visualized using Fiji/ImageJ 1.45r (National Institutes of Health) and analyzed for markers of apoptosis (MOMP, rounding, blebbing, chromatin condensation) either by eye or using MATLAB (MathWorks).
Mice
Nu/nu mice (Cox-7, Massachusetts General Hospital) were housed and handled according to Massachusetts General Hospital Institutional Animal Care and Use Committee guidelines. For procedures involving window chamber and tumor implantation, intravenous injection, and intravital imaging, mice were anesthetized by isoflurane inhalation at a flow rate of 2 L/minute isoflurane : 2 L/minute oxygen. For extended anesthesia, the isoflurane flow rate was gradually reduced to 1-2 L/minute. All surgical procedures were performed in a sterile surgery room using a stereomicroscope. For mice implanted with window chambers, antibiotics were administered via the drinking water. Anesthetized mice were monitored at all times. Titanium dorsal skin fold chambers (APJ Trading Co, Inc.) were surgically implanted in the dorsal skin fold of Nu/Nu mice, as described previously[1]. Tumor cells (1:1 mixture of reporter to non-fluorescent parent cell) in a 1:4 mixture with Matrigel (Becton Dickinson) were injected subcutaneously into the window of the chamber, and allowed to grow for approximately one week. The skin was removed and replaced with a 10 mm-diameter glass coverslip, secured in place with a C-clip. For intravital experiments, mice were treated with 100 mg/kg ABT-263 (12.5% DMSO/PEG-400, 100 μL total volume) i.p. once per day for 2 days.
Intravital microscopy
Anesthetized mice were positioned on a stable platform heated to 37°C with the dorsal window chamber mounted coverslip side facing up. Fluorescein isothiocyanate (FITC)-dextran (Sigma Aldrich), Angiosense 680/750 (Perkin Elmer, Inc), Annexin-Vivo 750 (Perkin Elmer, Inc.), or CLIO VT680 were administered via tail-vein catheter.
Tumors were imaged in vivo using a customized Olympus FV1000 BX61-W1 (Olympus America) confocal/multiphoton microscope with a programmable, motorized X-Y-Z stage. The objectives used were 2× /340 XLFluor (NA=0.14, air), 20× UPlanFL (NA=0.50, water), and 60× LUMFL N (NA=1.10, water). AcGFP1 and mCherry were excited using the 488nm line of an argon ion laser and a 559nm diode laser, respectively. Emitted light was separated using a 405/488/559/635nm dichroic beam splitter and the signal refined by the excitation band pass filters BA505-540 and BA575-605, respectively.
Annexin Vivo was visualized using the Olympus FV1000 and an Olympus IV100 fixed pinhole confocal microscope (Olympus America). The objectives used were the same as those for the FV1000 BX61- W1, and the lasers used to excite H2B-GFP and Annexin Vivo were 488/750nm, respectively. The emitted light was then filtered using a 560/640/750 dichroic mirror and refined with BA505-540 (GFP) and BA770IF (Annexin Vivo) longpass filters. Each of the signals was captured separately to minimize bleedthrough of fluorescent signals into neighboring channels. All images were stored and analyzed as 512 × 512 pixel (0.46mm × 0.46mm) 16-bit images.
Image processing and analysis
Microscopy images from in vivo or in vitro experiments were processed using collaborative digital filtering[10], and alpha-rooting to simultaneously reduce noise and enhance image details. More details are reported in the supplemental section.
Analysis of time-lapse images was performed using MATLAB (Mathworks) and Fiji/ImageJ 1.45r (National Institutes of Health). Only unprocessed images were used for image analysis. Image analysis for automated MOMP detection was done on individual frames of time-lapse image stacks. Cell detection and segmentation were initially performed using the brightfield images, from which segmentation masks around cells of interest could be created. Regions of interest (ROI) were then automatically selected within the cells, excluding areas of high intensity signal (from mitochondria), to detect subtle changes in cytosolic intensity. Similarly sized ROIs were also selected from the background. Median intensity values for both cytosol and background ROIs were determined and normalized by their areas; background signal was then subtracted from the cytosol signal. From the plotted data, a sharp change in slope is clearly seen over time; this change corresponds to the point at which MOMP occurs.
Results
Characterization of cell lines
IMS reporter protein (IMS-RP) is a genetic reporter of apoptosis that was created by fusing an FP to the first 59 N-terminal amino acids of the mitochondrial protein Smac/DIABLO[2, 9]. During apoptosis-associated MOMP, the FP is released from the IMS into the cytosol. While IMS-RP contains the mitochondrial targeting signal peptide, it does not contain the inhibitor of apoptosis protein (IAP) binding motif and is therefore biologically neutral. In cells transduced with IMS-RP green fluorescent protein (GFP), fluorescence was observed within the mitochondria (Figs. 1 and 2), co-localizing with both full-length endogenous Smac/DIABLO as well as with apoptosis inducing factor (AIF), another protein that resides within the mitochondrial IMS (Fig. S1). MDA-MB-231 cells typically showed filamentous mitochondria, which localized most abundantly around the nucleus (Fig. 1A). In contrast, PANC-1 cells typically contained shorter and more rounded mitochondria that were scattered throughout the entire cell (Fig. 2A). These mitochondrial phenotypes were consistent throughout all of our experiments, suggesting that the mitochondria of MDA-MB-231 and PANC-1 cells are morphologically distinct. In addition, IMS-RP fluorescence is brighter in the MDA-MB-231 cells, regardless of whether cells are transfected or transduced, which could influence the resultant images. Occasionally, round “swollen” mitochondria were seen in both cell lines, but the presence of these mitochondria did not correlate with apoptosis. In fact, recent findings suggest ill correlation between mitochondrial morphology and apoptosis, such that swollen or fragmented mitochondria do not predict apoptosis[12].
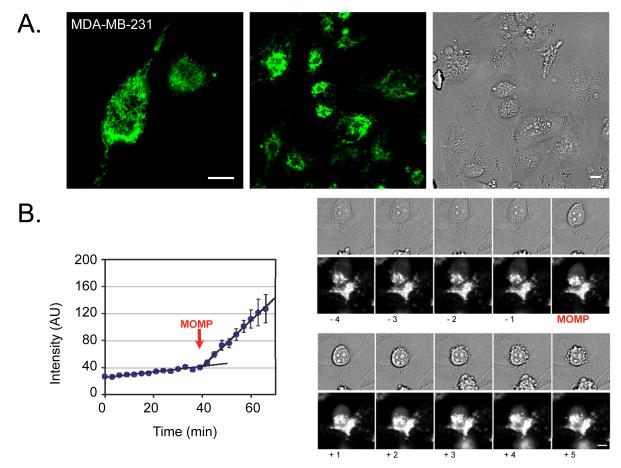
Fig. 1. MDA-MB-231 IMS-GFP cells in vitro.
A. MDA-MB-231 breast cancer cells at high (180x) and low (60x) magnification showing intermembrane space-green fluorescent protein (IMS-GFP) localized within the mitochondrial IMS. The panel on the far right shows the corresponding brightfield image of cells at low magnification. B. The image sequence on the right shows a cell undergoing mitochondrial outer membrane permeabilization (MOMP), rounding, and death over time, following treatment with ABT-263. The graph on the left shows the change in signal intensity for a region of interest in an apoptotic cell, both before and after undergoing MOMP (indicated by the red arrow). Error bars show SEM for multiple ROI within the cell over time. Scale bars are 10 μm.
Fig. 2. PANC-1 IMS-GFP cells in vitro.
A. PANC-1 pancreatic cancer cells at high (180x) and low (60x) magnification show IMS-GFP localized in the mitochondrial IMS. The panel on the far right shows the corresponding brightfield image of the cells at low magnification. B. The image sequence on the right shows a cell undergoing MOMP, rounding, and death over time, following drug treatment. The graph on the left shows the change in signal intensity of a region of interest in an apoptotic cell, both before and after undergoing MOMP (indicated by the red arrow). Error bars show SEM for multiple ROI within the cell over time. Note similar kinetics as in Fig. 1. Scale bars are 10 μm.
In cell culture, apoptosis occurred in ~0.6 % of MDA-MB-231 cells and in ~1.4 % of PANC-1 cells. Fig. 1B summarizes a typical and representative time series both before and after MOMP. The morphological changes (disappearance of distinct intracellular foci/appearance of diffuse cytosolic fluorescence) were readily visible, and were used to identify MOMP via image analysis algorithm (Figs. 1B and 2B). The sequence of events leading up to irreversible cell death (MOMP, rounding, blebbing, nuclear fragmentation) was defined for both cell lines. However, as previously observed in HeLa cells[13], biological variability was prevalent throughout the cell population. To better compare time scales of events and to provide statistics, we quantified these cell processes in real-time. For MDA-MB-231, ABT-263-induced MOMP at 135±85 minutes (mean ±SD), rounding occurred at 61±83 minutes, blebbing at 134±77 minutes, and chromatin condensation at 173±67 minutes; for PANC-1 cells, MOMP occurred at 363±37 minutes, rounding at 291±131 minutes, and blebbing at 386±83 minutes.
Characterization of in vivo tumors
While image analysis of sparsely plated cells in culture was relatively simple, in vivo analysis proved more complex due to cellular crowding. To enable easier identification of individual cells, a second fluorescent marker was introduced. For this, we selected H2B-FP, a nuclear histone protein, which when fused to an FP can be used to identify chromatin condensation, a later step in the apoptotic pathway[1]. We subsequently determined whether these tumors could grow in vivo, and if so how their growth and apoptotic rates compared to those in the in vitro environment.
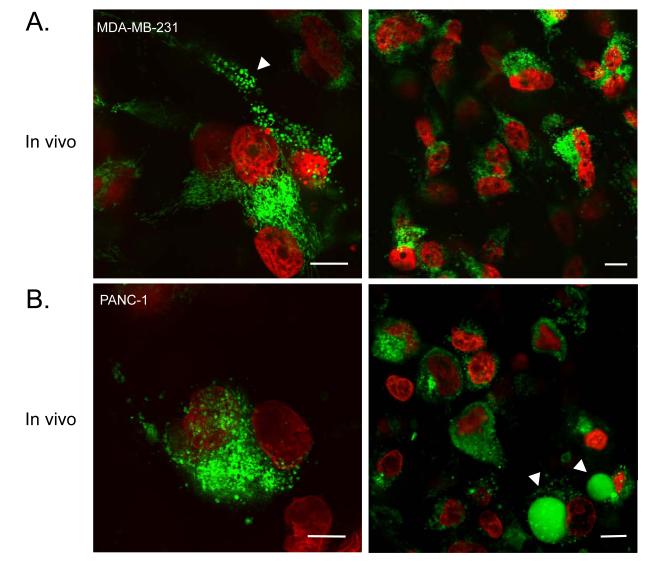
Both MDA-MB-231-IMS/H2B and PANC-1-IMS/H2B cells grew well in xenografts, demonstrating similar growth characteristics to the parental cell lines (Fig. 3). Both cell lines also grew readily in window chambers. At 60x magnification, individual mitochondria could be easily discerned within individual cells in vivo (Fig. 3). We were thus able to verify that mitochondria have a similar appearance in vivo as in vitro. Furthermore, cells undergoing apoptosis in vivo could be easily identified using the dual fluorescent marker. One significant difference between our in vitro and in vivo measurements, however, was in the basal apoptosis rates of cells. For both cell lines tested, we observed higher apoptosis rates in vivo than in vitro (MDA-MB-231-IMS/H2B: 0.6% in vitro versus 11.6% in vivo; PANC-1-IMS/H2B: 1.4% in vitro versus 8.6% in vivo). These differences could be explained by the presence of selective pressures in the tumor microenvironment, which are not encountered in culture conditions.
Fig. 3. In vivo images of breast (top) or pancreatic (bottom) cancer cells expressing IMS and H2B reporter proteins in a live mouse.
A. MDA-MB-231 cancer cells in vivo at high (left) and low (right) magnification. The white arrow points to a cell containing swollen mitochondria. B. PANC-1 cancer cells in vivo at high (left) and low (right) magnification. Arrows point to apoptotic blebs. Note the cellular detail achievable by in vivo imaging. Scale bars are 10 μm.
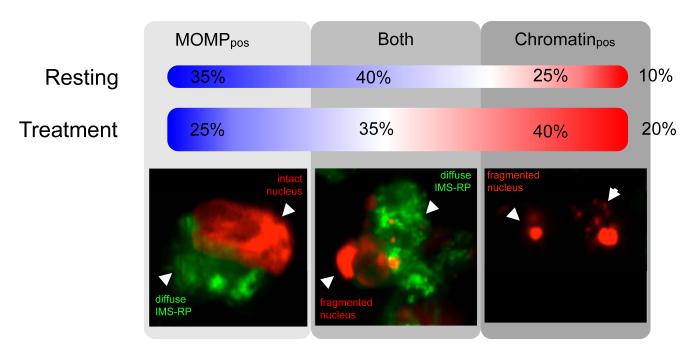
The availability and use of both MOMP and chromatin condensation/fragmentation reporters was useful not only for identifying individual cells but also for temporarily assessing the apoptotic process (Fig. 4). Under basal state conditions (~10% of cells in apoptosis), 35% of apoptotic cells were MOMP positive only, 25% only showed chromatin condensation, and 40% underwent both MOMP and chromatin condensation. In drug treated tumors (~20% of cells in apoptosis), apoptotic cells were seen to shift slightly in favor of chromatin condensation states.
Fig. 4. Percentage of apoptotic cells undergoing MOMP and/or chromatin condensation in treated and untreated tumors.
Apoptosis typically involves caspase activation followed by MOMP and subsequent chromatin condensation. In vivo, cells are also then cleared by phagocytic cells. The percentage of apoptotic cells undergoing MOMP and/or chromatin condensation in treated and untreated tumors (n=3 for each cell type) was assessed qualitatively. Images show examples of cells at each stage. The percentages show the number of cells under resting (~10% apoptosis) and ABT-263 treatment conditions (~20% apoptosis), and their respective distributions in the apoptotic cascade. MOMPpos refers to cells undergoing MOMP only and Chromatinpos refers to chromatin condensed/fragmented cells only. Observations in the treatment group were carried approximately one day after i.p. injection of ABT-263.
Measuring drug induced apoptosis at the single cell level in vivo
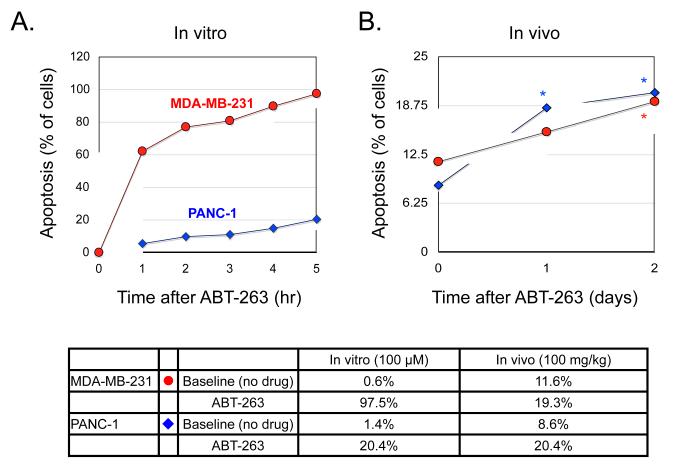
We next determined whether drug treatment effects could be monitored directly in vivo at the single cell level (“single cell pharmacodynamics”). To do this, we treated established tumors with the Bcl-2 inhibitor, navitoclax (ABT-263), a drug currently undergoing clinical trials both as a combination treatment (with Docetaxel, Paclitaxel, Erlotinib, Rituximab, Gemcitabine, among others) and as a monotherapy. In parallel, and as a comparison, we performed similar experiments in cell culture. We found that while a single dose of navitoclax (100 μM) in vitro induced apoptosis in nearly all MDA-MB-231-IMS/H2B cells within a 5 hour period, it only induced apoptosis in ~20% of PANC-1-IMS/H2B cells under the same conditions (Fig. 5). These results are consistent with earlier findings, which showed that while EC50 values for navitoclax ranged from 0.11-39 μM in small cell lung cancer and hematologic cancer lines, most other solid tumors are not as responsive to navitoclax treatment alone[14, 15].
Fig. 5. Comparison of ABT-263 kinetics in vitro and in vivo.
A. MDA-MB-231 and PANC-1 cells were treated with 100 μM ABT-263 and then imaged for 5 hours. Images were analyzed for the percentage of cell death occurring in the population. Graphs show data from three separate experiments per cell line. B. MDA-MB-231 and PANC-1 tumors growing in window chambers in nude mice (n=3 per line) were imaged before treatment, 1 day and 2 days after treatment with ABT-263 (100 mg/kg i.p.). The tumors were then analyzed for the percentage of cell death occurring within the population. Statistical significance, indicated by asterisks, was determined using a two-tailed unpaired Student’s t test (PANC-1 p<0.01, MDA-MB-231 p<0.02). The table below compares the percentage of cells undergoing apoptosis in culture versus in mice, with and without drug treatment at the specified times.
When tested in vivo, drug induced apoptosis was readily identifiable. In order to approximately match the drug dose encountered by cells in vitro[16], 100 mg/kg of navitoclax was injected intraperitoneally (i.p.) into mice. Previous studies have determined this dose to be efficacious without inducing major thrombocytopenia[14, 17]. While the pancreatic and breast cancer cell lines showed divergent behavior in vitro, apoptosis rates of the cell lines in vivo were remarkably similar. Furthermore, apoptosis rates in vivo plateaued at ~20%, a 100% increase from basal levels. Our studies therefore indicate that both tumor types respond to navitoclax monotherapy in vivo. Apoptotic cells did not demonstrate any preferential distribution, either in their distance from the tumor microvessels or within the tumors themselves.
Comparison with exogenous stains
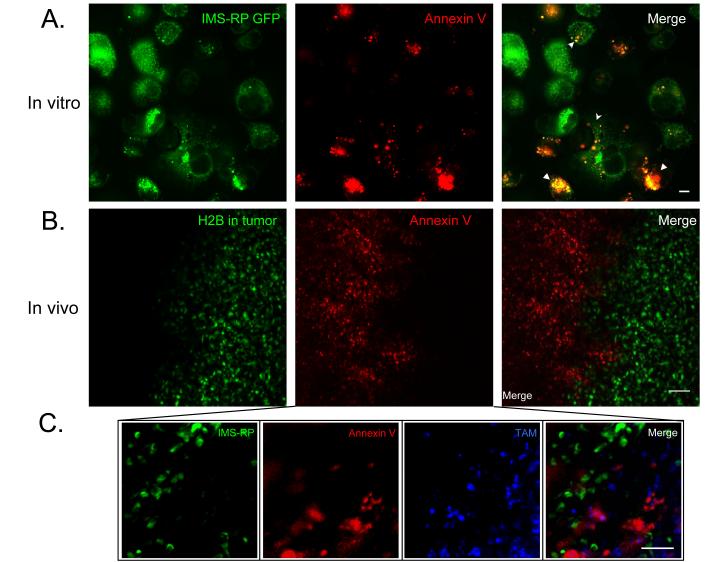
Drug induced apoptosis is often measured via non-genetic means using fluorescent Annexin V or other affinity ligands (e.g. NucView). While the Annexin V approach is particularly well established in vitro, its validity in in vivo settings has not yet been explored at the single cell level. We resolved to determine whether systemic injection of Annexin V in mice would result in its accumulation in apoptotic cells (defined by MOMP and chromosome condensation). Interestingly, we observed that nearly all fluorescently labeled Annexin V co-localized with tumor associated macrophages (TAM) outside of the tumor rather than with apoptotic tumor cells (Fig. 6). Indeed, TUNEL-stained apoptotic cells failed to show significant colocalization with Annexin V in vivo (Fig. S3). We hypothesize that dying cancer cells recruit monocytes to the tumor site, where they differentiate into TAM, and result in both cell types accumulating the imaging agent. Assessment of TAM may therefore represent an indirect means for identifying cell death for some tumor types, reporting on the macrophage response rather than on apoptosis directly.
Fig. 6. Annexin V stains apoptotic cells in vitro, and macrophages in vivo.
A. PANC-1 IMS-RP cells grown in cell culture were treated with drug and Annexin Vivo750, before undergoing imaging 3 hours later. Annexin V staining was seen to increase in cells undergoing apoptosis in the vitro setting. Scale bar are 10 μm. B. Mice with PANC-1 IMS/H2B tumors growing in window chambers were injected intravenously with Annexin Vivo, and imaged within 1 hour post-injection. The Annexin Vivo primarily stained the area surrounding the tumor, rather than in apoptotic tumor cells. C. Higher resolution imaging at the edge of an MDA-MB-231 tumor shows Annexin Vivo 750 to colocalize outside of the tumor with tumor associated macrophages (stained by CLIO-VT680) rather than apoptotic tumor cells. Scale bars are 50 μm.
Discussion
The use of high spatial resolution imaging has become widespread in cell biology. However, the attainment of similar resolutions in vivo has remained challenging due to motion, limited achievable signal-to-noise ratios and restricted tissue penetration of light [18]. In view of the findings showing that many cellular processes and responses can behave differently in vivo compared to in vitro, it is now clear that the ability to image at subcellular resolution in vivo would have important consequences. Here, we not only show that it is practically feasible to image apoptotic events within individual cells over time, but also that it is possible to measure drug induced effects in the living animal. These results are important for several reasons. Firstly, such technological advances will likely broaden the study of apoptosis and its regulation at both the single cell and population level in vivo. Secondly, the use of such a method could be applied as a tool in systems pharmacology for measuring the therapeutic effect (or failure) of drugs on individual cells, cell populations or cell niches. Thirdly, if and when combined with fluorescent drugs[19, 20], this approach could serve to improve our ability to map both the pharmacokinetics and pharmacodynamics of drugs. Similarly, the toxicity of drugs on non-malignant cells could be determined in transgenic mice that express the reporters. Finally, our findings suggest the possibility that other cellular structures and pathways could likewise be imaged at high spatial resolutions in vivo.
Several studies have used intravital microscopy to study tumor immunology [21-23], microvascularity[24], invasion and metastases[25-27], as well as cell cycle progression[1, 18]. Many of these studies, however, used lower resolutions to visualize cell-cell interactions or cellular movements over time. In our investigation, because our aim was to resolve intracellular details (not generally needed for cell tracking studies), we required much higher spatial resolutions. We achieved this by creating highly fluorescent dual reporter lines and then carefully optimizing the acquisition conditions. IMS-RP is a biologically neutral reporter that lacks the IAP binding motif. As a result, there are no biological side-effects from over-expression of the Smac/DIABLO peptide (as there would from expression of full length proteins fused to FPs). Lentiviral constructs provide a rapid method for labeling cells, including primary cells. A major technical challenge in intravital microscopy, however, is overcoming artifacts caused by physiological motion. In our study, to reduce motion during prolonged imaging sessions, the window chambers of isoflurane-anesthetized mice were immobilized on a heated stage, and the mice were monitored carefully throughout each experiment. Scan speeds were likewise adjusted to reduce artifacts. To obtain a three-dimensional representation of the field of view, all images were collected as z-stacks using a high magnification water-immersion objective.
Despite numerous technological advances in intravital imaging, there remain a number of unresolved practical issues. Firstly, to obtain the highest possible spatial resolution, capable of resolving mitochondria and chromatin within cells, the present study required the use of window chamber models. In future research, it would be interesting to extend this type of imaging to the study of orthotopic cancers in genetically engineered mouse models. For this to be possible, however, better immobilization strategies and optics are needed. We nevertheless remain confident that with continuing advances in miniaturized optics[28, 29], fast scanning and newer immobilization techniques, such studies will ultimately be feasible. A second area in need of improvement is in the design of reporter constructs. For example, the use of more red shifted[30] or brighter FPs would increase the signal-to-noise ratio and thus facilitate image analysis. Alternatively, the creation and use of turn-on FP reporters that only fluoresce during apoptosis would be another interesting means of obtaining information about the signaling pathways involved in this process. Several caspase-activatable probes have recently been described for detection of cell death in vitro[3]; but these have yet to be tested in vivo. Another area demanding significant attention is in the development of automated image analysis and quantitation, particularly for use in unsupervised paradigms[31-33]. Finally, there is an obvious need for deeper imaging into tumors. It is possible that this issue could be addressed through the use of multiphoton approaches, more red shifted constructs and perhaps newer illumination schemes[34].
The in vivo studies presented here not only complement earlier investigations in HeLa cells[2, 13, 35], but also serve to further validate these studies in breast and pancreatic cancer cell lines. Several important differences should nevertheless be highlighted. For example, Spencer et al. primarily used a combination of TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) and cycloheximide treatment to induce cell death. In cases where TRAIL was used alone, however, it appears the researchers required five-fold greater concentrations of TRAIL to induce cell death, which is comparable to the concentration used in our study to induce cell death within a similar time frame. In addition, we also found that the duration of MOMP in both PANC-1 and MDA-MB-231 cells was similar to that seen in HeLa cells (for both, it occurred in minutes). In the present study, we used navitoclax rather than TRAIL to induce apoptosis on the basis that this drug is currently under investigation in clinical trials. Navitoclax (ABT-263) is a small molecule inhibitor of the pro-survival Bcl-2 family of proteins, which include Bcl-2, Bcl-xL, and Mcl-1. From studies of small-cell lung carcinoma and acute lymphoblastic leukemia, the reported effective dose range for navitoclax is approximately 110 nM to 39 μM[17]. In our own experiments, while 25 μM ABT-263 was sufficient to cause cell death in approximately 50% of MDA-MB-231 cells, it did not cause significant apoptosis in PANC-1 cells. We thus increased the in vitro dose of ABT-263 to 100 μM.
In summary, we believe that by continuing to make advances in microscopic imaging and apoptotic reporter constructs, these tools could soon have broad applications for biology and drug development. We expect that the use of IMS-RP, together with other genetic reporters, will not only allow us to visualize multiple components of the apoptotic signaling pathways (which will likely differ depending on drug treatment/cell type), but will also be useful for drug efficacy profiling. Naturally, such reporters will need to be applied to the study of other forms of drug-induced cell death, e.g. by the creation of in vivo reporters for DNA damage[36], senescence[4], autophagy[5], and necrosis, among others[37].
Supplementary Material
Acknowledgments
We would like to thank Drs. Kohler, Sergeyev, Fisher-Jeffes, Thurber, Yoshiko Iwamoto, and all members of the Pittet Lab for valuable technical suggestions, review and reagents. Our special thanks go to our collaborators for their invaluable discussions: Drs. Timothy Mitchison (H2B constructs), Peter Sorger (IMS-FP constructs), Mikael Pittet, Anthony Letai, Gaudenz Danuser and Charles Lin. This work was supported in part by NIH grants: 2P50CA086355, 2R01EB006432, 1P01CA139980 and 1R01CA164448.
References
- 1.Orth JD, Kohler RH, Foijer F, Sorger PK, Weissleder R, Mitchison TJ. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71:4608–16. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholls SB, Chu J, Abbruzzese G, Tremblay KD, Hardy JA. Mechanism of a genetically encoded dark-to-bright reporter for caspase activity. J Biol Chem. 2011;286:24977–86. doi: 10.1074/jbc.M111.221648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian F, Deguchi K, Yamashita T, et al. In vivo imaging of autophagy in a mouse stroke model. Autophagy. 2010;6:1107–14. doi: 10.4161/auto.6.8.13427. [DOI] [PubMed] [Google Scholar]
- 6.Ntziachristos V, Schellenberger EA, Ripoll J, et al. Visualization of antitumor treatment by means of fluorescence molecular tomography with an annexin V-Cy5.5 conjugate. Proc Natl Acad Sci U S A. 2004;101:12294–9. doi: 10.1073/pnas.0401137101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankenberg FG, Vanderheyden JL, Strauss HW, Tait JF. Radiolabeling of HYNIC-annexin V with technetium-99m for in vivo imaging of apoptosis. Nat Protoc. 2006;1:108–10. doi: 10.1038/nprot.2006.17. [DOI] [PubMed] [Google Scholar]
- 8.Zhao M, Beauregard DA, Loizou L, Davletov B, Brindle KM. Non-invasive detection of apoptosis using magnetic resonance imaging and a targeted contrast agent. Nat Med. 2001;7:1241–4. doi: 10.1038/nm1101-1241. [DOI] [PubMed] [Google Scholar]
- 9.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 10.Dabov K, Foi A, Katkovnik V, Egiazarian K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans Image Process. 2007;16:2080–95. doi: 10.1109/tip.2007.901238. [DOI] [PubMed] [Google Scholar]
- 11.Fumene Feruglio P, Vinegoni C, Gros J, Sbarbati A, Weissleder R. Block matching 3D random noise filtering for absorption optical projection tomography. Phys Med Biol. 2010;55:5401–15. doi: 10.1088/0031-9155/55/18/009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reis Y, Bernardo-Faura M, Richter D, et al. Multi-Parametric Analysis and Modeling of Relationships between Mitochondrial Morphology and Apoptosis. PLoS One. 2012;7:e28694. doi: 10.1371/journal.pone.0028694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–32. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse C, Shoemaker AR, Adickes J, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–8. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 15.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 16.Park CM, Bruncko M, Adickes J, et al. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51:6902–15. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 17.Shoemaker AR, Mitten MJ, Adickes J, et al. Activity of the Bcl-2 family inhibitor ABT-263 in a panel of small cell lung cancer xenograft models. Clin Cancer Res. 2008;14:3268–77. doi: 10.1158/1078-0432.CCR-07-4622. [DOI] [PubMed] [Google Scholar]
- 18.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–91. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiner T, Earley S, Turetsky A, Weissleder R. Bioorthogonal small-molecule ligands for PARP1 imaging in living cells. Chembiochem. 2010;11:2374–7. doi: 10.1002/cbic.201000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budin G, Yang KS, Reiner T, Weissleder R. Bioorthogonal Probes for Polo-like Kinase 1 Imaging and Quantification. Angew Chem Int Ed Engl. 2011;50:9378–81. doi: 10.1002/anie.201103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iannacone M, Moseman EA, Tonti E, et al. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 2010;465:1079–83. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrickson SE, Mempel TR, Mazo IB, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol. 2008;9:282–91. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celli S, Albert ML, Bousso P. Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat Med. 2011;17:744–9. doi: 10.1038/nm.2376. [DOI] [PubMed] [Google Scholar]
- 24.Vakoc BJ, Lanning RM, Tyrrell JA, et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15:1219–23. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Entenberg D, Wyckoff J, Gligorijevic B, et al. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6:1500–20. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–87. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–57. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 28.Alencar H, Mahmood U, Kawano Y, Hirata T, Weissleder R. Novel multiwavelength microscopic scanner for mouse imaging. Neoplasia. 2005;7:977–83. doi: 10.1593/neo.05376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barretto RP, Ko TH, Jung JC, et al. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nat Med. 2011;17:223–8. doi: 10.1038/nm.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaqaman K, King EM, Amaro AC, et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J Cell Biol. 2010;188:665–79. doi: 10.1083/jcb.200909005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoma CR, Matov A, Gutbrodt KL, et al. Quantitative image analysis identifies pVHL as a key regulator of microtubule dynamic instability. J Cell Biol. 2010;190:991–1003. doi: 10.1083/jcb.201006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaqaman K, Loerke D, Mettlen M, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andresen V, Alexander S, Heupel WM, Hirschberg M, Hoffman RM, Friedl P. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr Opin Biotechnol. 2009;20:54–62. doi: 10.1016/j.copbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Rehm M, Huber HJ, Hellwig CT, Anguissola S, Dussmann H, Prehn JH. Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ. 2009;16:613–23. doi: 10.1038/cdd.2008.187. [DOI] [PubMed] [Google Scholar]
- 36.Fan JY, Cui ZQ, Wei HP, et al. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem Biophys Res Commun. 2008;367:47–53. doi: 10.1016/j.bbrc.2007.12.101. [DOI] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.