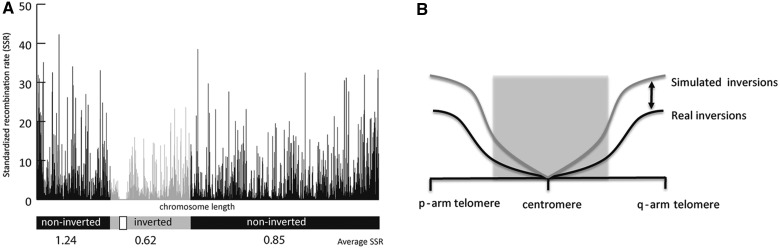
Fig. 3.
Distribution of recombination rates. (A) SRR along the human chromosome 4. SRR (y axis) are shown as needles across nonoverlapping windows of 10 kb in the whole chromosomal length (x axis). The genomic region affected by an inversion is depicted in gray, whereas noninverted regions are showed in black. The white rectangle indicates the centromere. Average recombination rate for each region is shown in numbers in the x axis. (B) Schematic representation of how recombination rates (SRR) varies along observed and simulated inversions. Average SRR across inverted and noninverted regions in observed (black) and simulated (dark gray) chromosomes. The light gray square shows the inverted region around the centromere. Differences between simulated and observed inversions are significantly different (t test, P value < 0.0001) (see text for details).