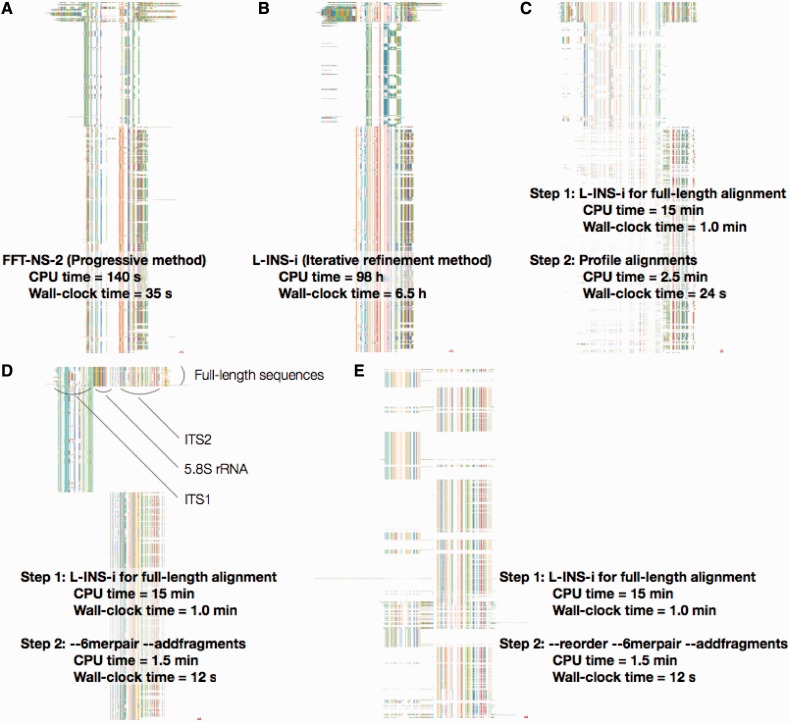
Fig. 2.
ITS alignments by different options of MAFFT, displayed on Jalview (Waterhouse et al. 2009). (A, B) Incorrect alignments by the FFT-NS-2 and L-INS-i algorithms, respectively. (C) An incorrect alignment by mafft-profile. The full-length sequences were aligned with the L-INS-i algorithm and then each new sequence was separately added to the full-length alignment, using mafft-profile. (D) Reasonable alignment by a two-step strategy. The ––6merpair ––addfragments option was used at the second step. (E) Reordered version of D; sequences are ordered such that similar sequences are placed closely. All calculations were performed using 16 cores on a Linux PC with 2.67 GHz Intel Xeon E7-8837/256 GB RAM.