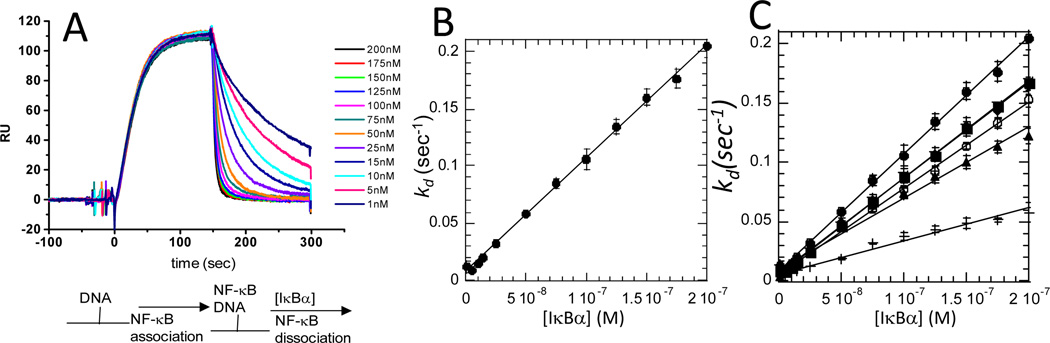
Figure 8.
(A) Real-time SPR binding experiment in which κB-site DNA was bound to the streptavidin chip at t=0, then NF-κB(p50(19-363)/p651-35) was allowed to associate with the DNA, and finally varying concentrations of IκBα were injected through the second sample loopand the dissociation rate constant (kd) was measured 41. A schematic of the binding events is shown below the graph. (B) Dissociation rate constants for active dissociation are plotted as a function of IκBα variant concentration. (C) Comparison of the active dissociation rate constants of IκBα folding variants: wild type IκBα (●), IκBα(C186P, A220P) (■), IκBα(Q111G) (▲), IκBα(Y254L/Q255H) (►), IκBα(Y254L/T257A) (◄) 41.