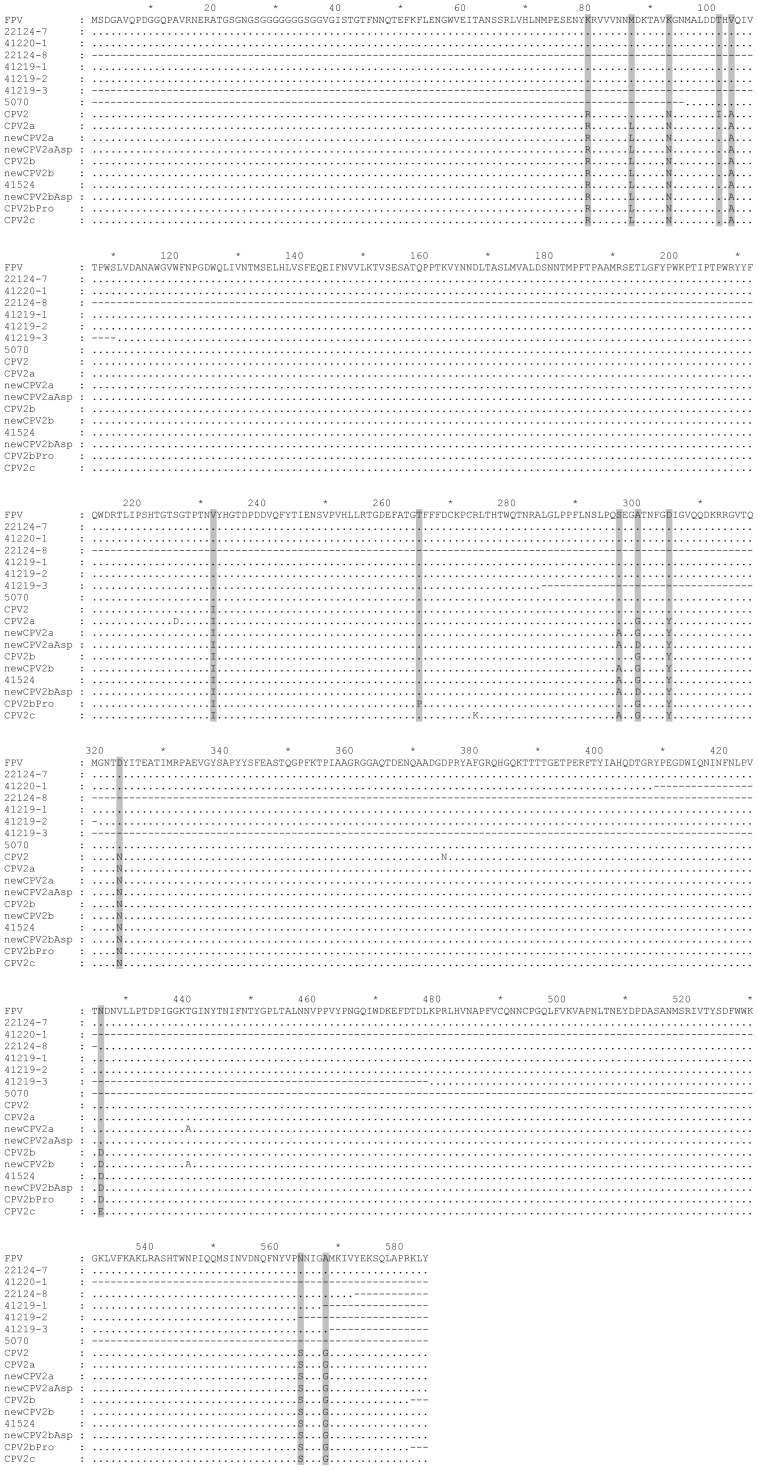
Figure 4. Multiple alignment of VP2 complete amino acid sequences.
CLUSTAL W was used to align the deduced VP2 sequences from wild carnivore parvoviruses characterized during this study and from parvoviruses representatives of each virus type. Characteristic amino acid positions are shaded. The amino acid sequences represented correspond to a lion isolate (EF418569), on the top, sequences 22124-7 (JF422105), 41220-1 and 22124-8 (JF422106) to E. mongooses, sequences 41219-1, 41219-2 and 41219-3 to red foxes, and sequence 41524 to the stone marten isolate. Other virus types are represented by strains M23255 (CPV-2), DQ340410 (CPV-2a), DQ340422 (newCPV-2a), AB054222 (newCPV-2a-Asp300), AF306450 (CPV-2b), AB054221 (newCPV-2b), AB054224 (newCPV-2b-Asp300), AF306449 (CPV2b-Pro265) and FJ005240 (CPV-2c).