Table 3. Fitting parameters in the duplication-divergence model for all organisms.
| Species |
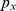
|
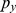
|
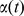
|
||||||||
| prNOG | roNOG | maNOG | veNOG | inNOG | meNOG | fuNOG | KOG/euNOG | COG/NOG | |||
| eco | 0.7 | 0.0008 | 0.007 | ||||||||
| sce | 0.7 | 0.0002 | 0.0008 | 0.0007 | 0.001 | ||||||
| ath | 0.7 | 0.0001 | 0.003 | 0.008 | |||||||
| cel | 0.5 | 0.0004 | 0.002 | 0.001 | 0.005 | ||||||
| dme | 0.5 | 0.0004 | 0.003 | 0.004 | 0.004 | 0.004 | |||||
| mmu | 0.7 | 0.0002 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.003 | |||
| hsa | 0.7 | 0.0002 | 0.0002 | 0.0004 | 0.0005 | 0.0005 | 0.0003 | 0.0004 | |||
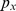 and
and 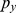 are time-independent and describe the probability that an interaction is retained after a duplication and the probability that an interaction is created de novo, respectively. The fraction of interacting pairs in the ancestral network at time
are time-independent and describe the probability that an interaction is retained after a duplication and the probability that an interaction is created de novo, respectively. The fraction of interacting pairs in the ancestral network at time  is represented by
is represented by 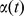 . There are in total nine ancestral time levels for the organisms investigated: the ancestral primates (prNOG), the ancestral rodents (roNOG), the ancestral mammals (maNOG), the ancestral vertebrates (veNOG), the ancestral insects (inNOG), the ancestral animals (meNOG), the ancestral fungi (fuNOG), the ancestral eukaryotes (KOG/euNOG), and the LUCA (COG/NOG). Existing time levels are specific for every species depending on its lineage.
. There are in total nine ancestral time levels for the organisms investigated: the ancestral primates (prNOG), the ancestral rodents (roNOG), the ancestral mammals (maNOG), the ancestral vertebrates (veNOG), the ancestral insects (inNOG), the ancestral animals (meNOG), the ancestral fungi (fuNOG), the ancestral eukaryotes (KOG/euNOG), and the LUCA (COG/NOG). Existing time levels are specific for every species depending on its lineage.
