Abstract
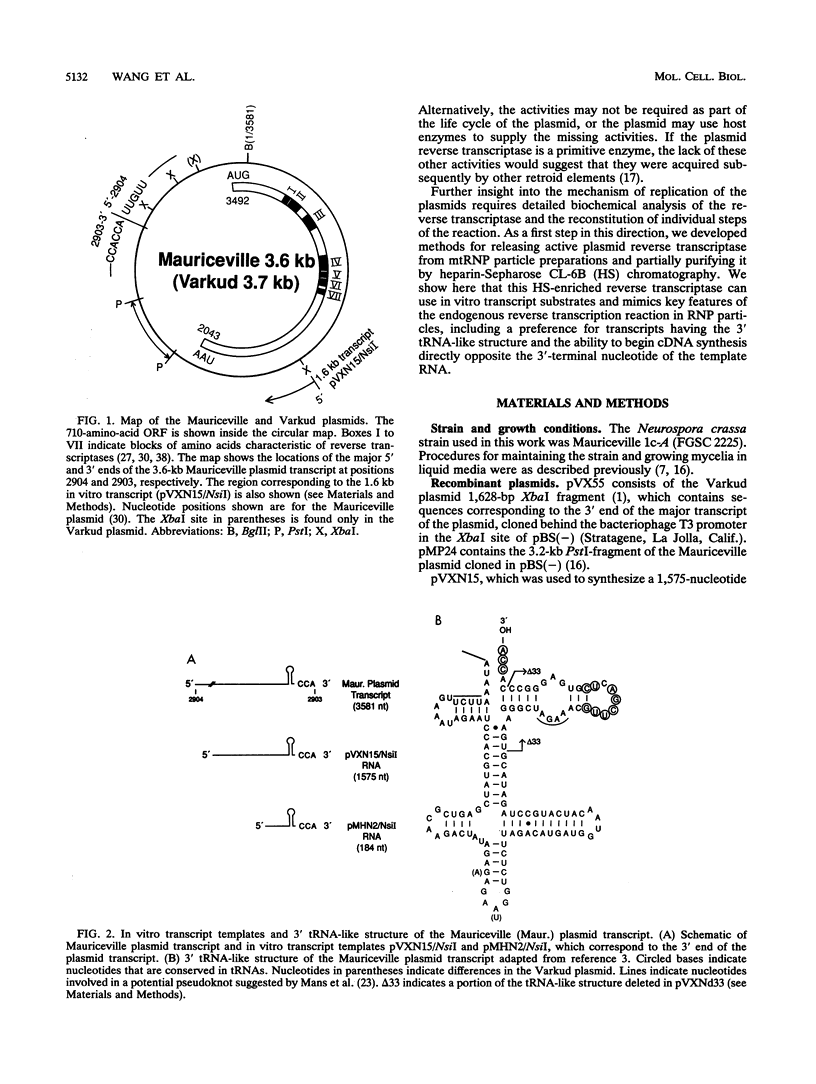
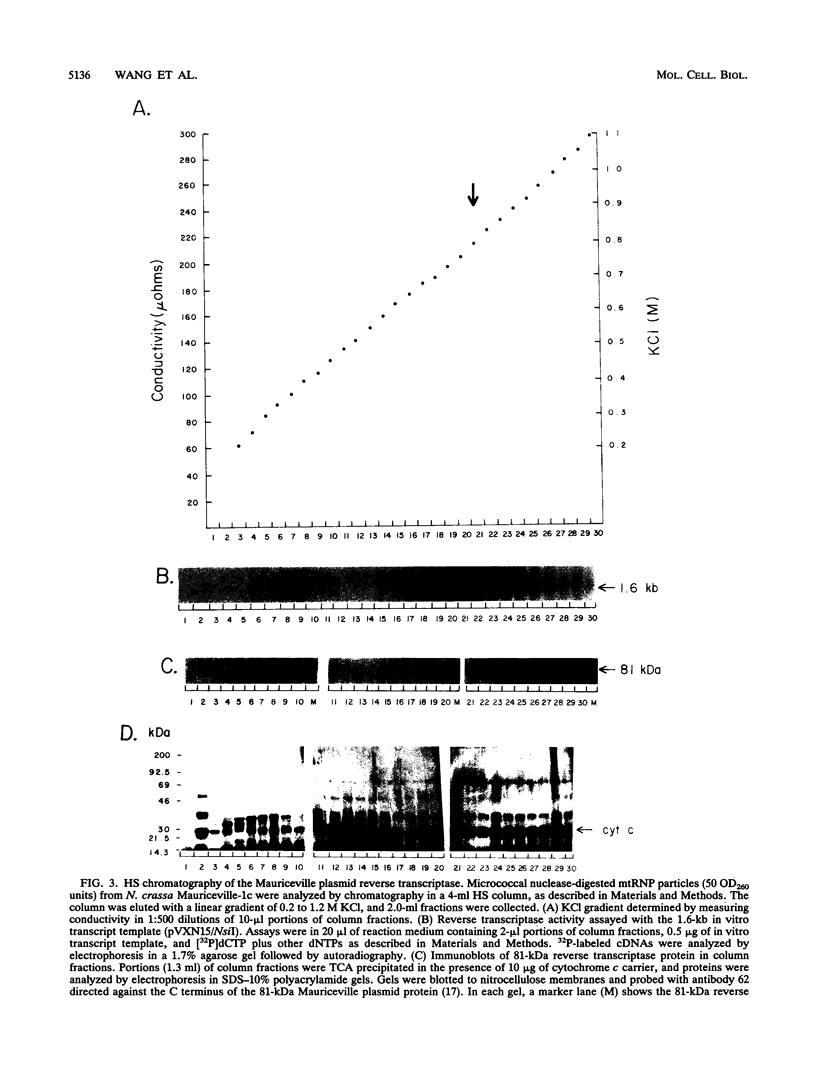
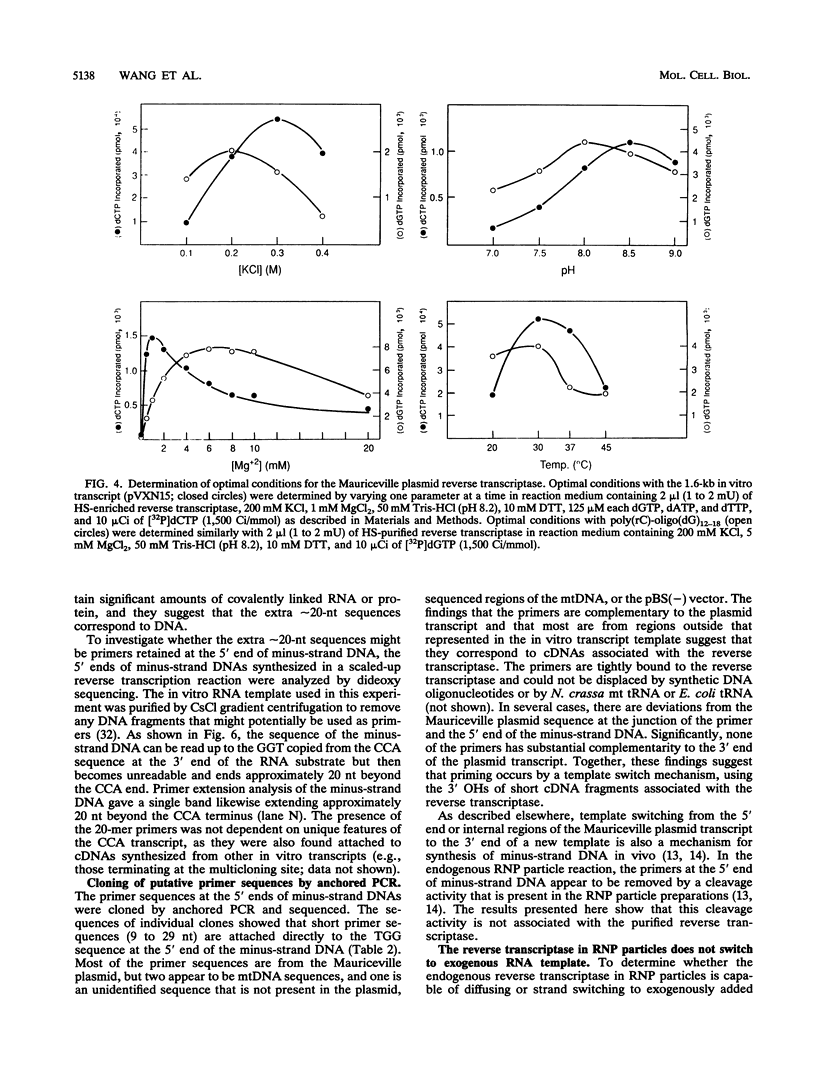
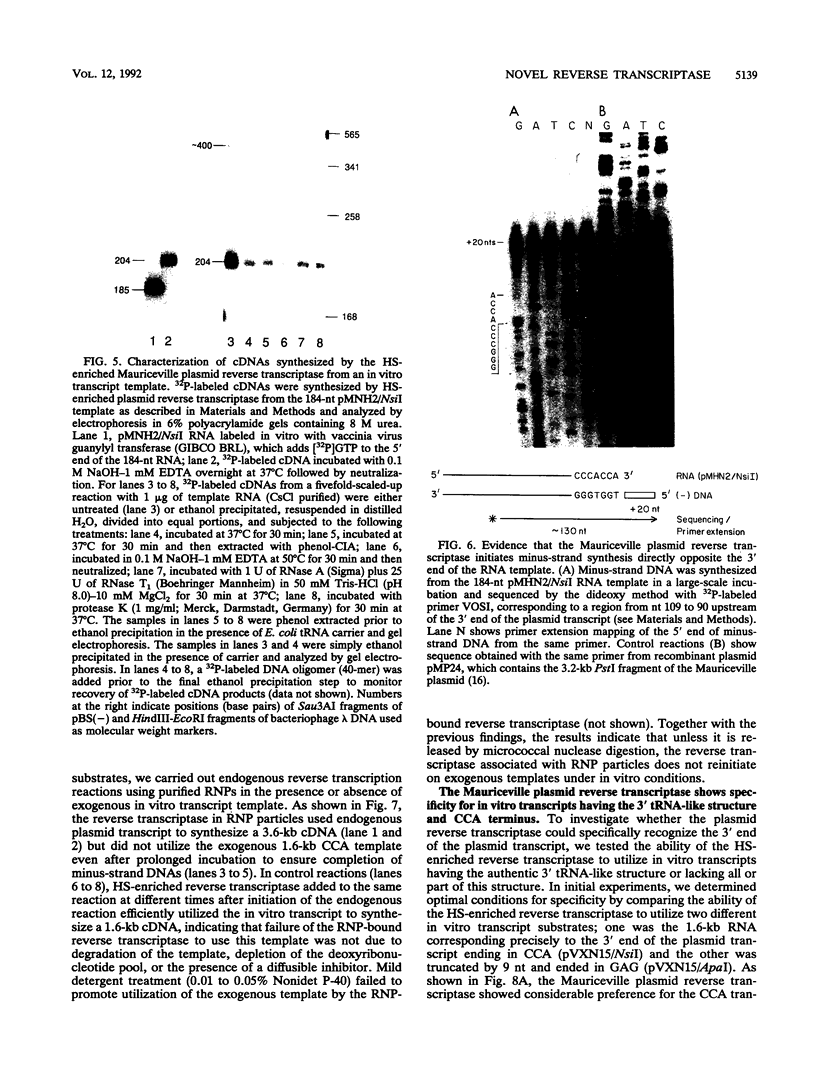
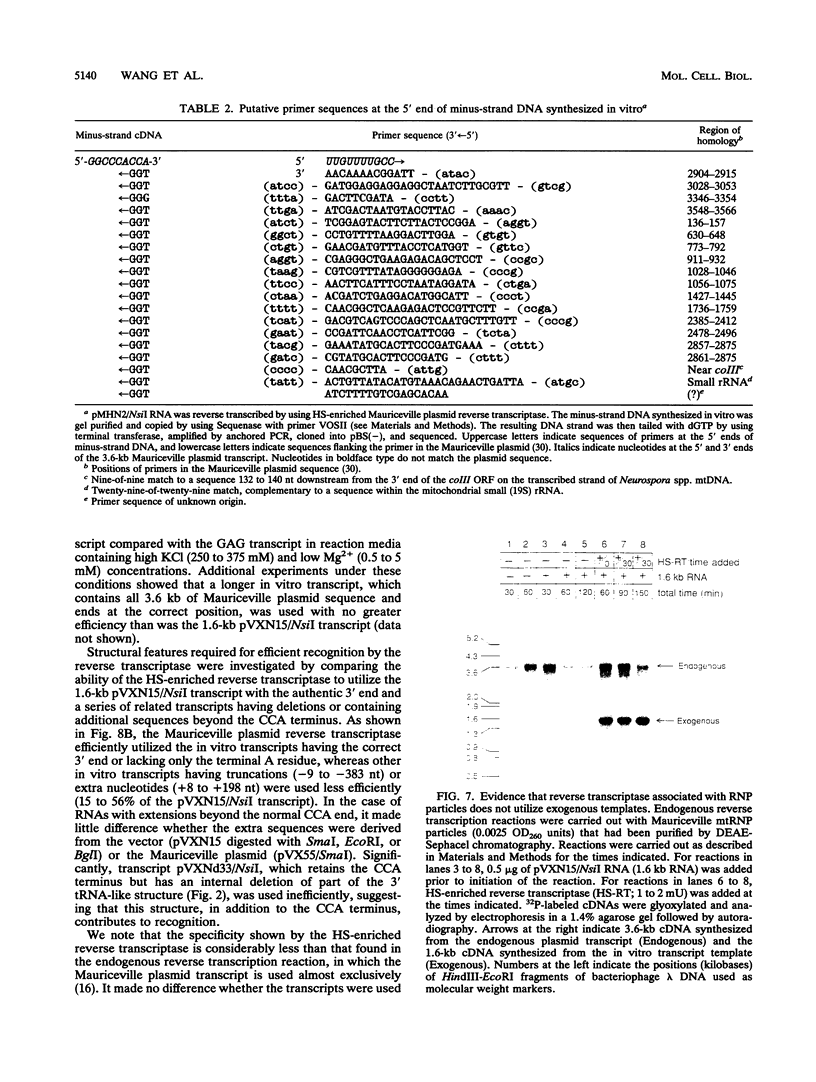
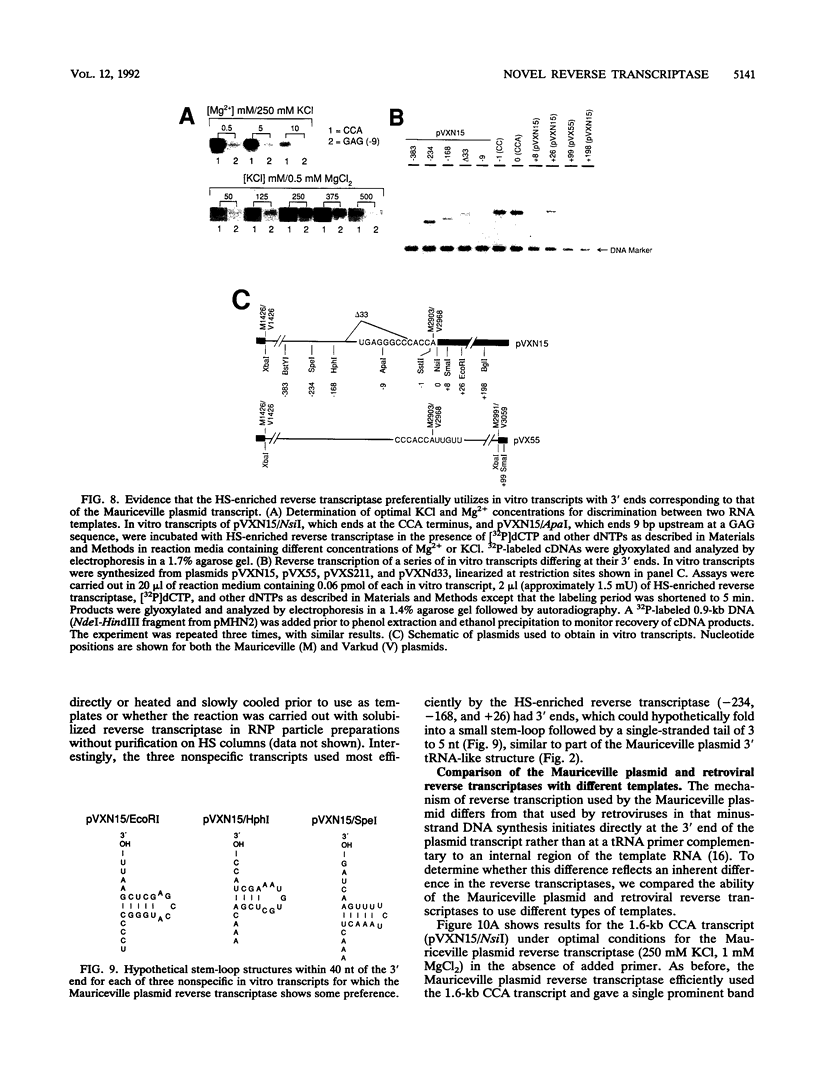
The Mauriceville and Varkud plasmids are retroid elements that propagate in the mitochondria of some Neurospora spp. strains. Previous studies of endogenous reactions in ribonucleoprotein particle preparations suggested that the plasmids use a novel mechanism of reverse transcription that involves synthesis of a full-length minus-strand DNA beginning at the 3' end of the plasmid transcript, which has a 3' tRNA-like structure (M. T. R. Kuiper and A. M. Lambowitz, Cell 55:693-704, 1988). In this study, we developed procedures for releasing the Mauriceville plasmid reverse transcriptase from mitochondrial ribonucleoprotein particles and partially purifying it by heparin-Sepharose chromatography. By using these soluble preparations, we show directly that the Mauriceville plasmid reverse transcriptase synthesizes full-length cDNA copies of in vitro transcripts beginning at the 3' end and has a preference for transcripts having the 3' tRNA-like structure. Further, unlike retroviral reverse transcriptases, the Mauriceville plasmid reverse transcriptase begins cDNA synthesis directly opposite the 3'-terminal nucleotide of the template RNA. The ability to initiate cDNA synthesis directly at the 3' end of template RNAs may also be relevant to the mechanisms of reverse transcription used by LINEs, group II introns, and other non-long terminal repeat retroid elements.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Grant D. M., Stohl L. L., Bottorff D. A., Nargang F. E., Lambowitz A. M. Nucleotide sequence of the Varkud mitochondrial plasmid of Neurospora and synthesis of a hybrid transcript with a 5' leader derived from mitochondrial RNA. J Mol Biol. 1988 Nov 5;204(1):1–25. doi: 10.1016/0022-2836(88)90594-3. [DOI] [PubMed] [Google Scholar]
- Akins R. A., Kelley R. L., Lambowitz A. M. Characterization of mutant mitochondrial plasmids of Neurospora spp. that have incorporated tRNAs by reverse transcription. Mol Cell Biol. 1989 Feb;9(2):678–691. doi: 10.1128/mcb.9.2.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins R. A., Kelley R. L., Lambowitz A. M. Mitochondrial plasmids of Neurospora: integration into mitochondrial DNA and evidence for reverse transcription in mitochondria. Cell. 1986 Nov 21;47(4):505–516. doi: 10.1016/0092-8674(86)90615-x. [DOI] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Corces V. G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Stohl L. L., Cole M. D., Lambowitz A. M. Characterization of a novel plasmid DNA found in mitochondria of N. crassa. Cell. 1981 May;24(2):443–452. doi: 10.1016/0092-8674(81)90335-4. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Hall T. C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988 May 5;201(1):31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Kuiper M. T., Lambowitz A. M. A novel reverse transcriptase activity associated with mitochondrial plasmids of Neurospora. Cell. 1988 Nov 18;55(4):693–704. doi: 10.1016/0092-8674(88)90228-0. [DOI] [PubMed] [Google Scholar]
- Kuiper M. T., Sabourin J. R., Lambowitz A. M. Identification of the reverse transcriptase encoded by the Mauriceville and Varkud mitochondrial plasmids of Neurospora. J Biol Chem. 1990 Apr 25;265(12):6936–6943. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M. Preparation and analysis of mitochondrial ribosomes. Methods Enzymol. 1979;59:421–433. doi: 10.1016/0076-6879(79)59103-4. [DOI] [PubMed] [Google Scholar]
- Lazcano A., Fastag J., Gariglio P., Ramírez C., Oró J. On the early evolution of RNA polymerase. J Mol Evol. 1988;27(4):365–376. doi: 10.1007/BF02101199. [DOI] [PubMed] [Google Scholar]
- Loh E. Y., Elliott J. F., Cwirla S., Lanier L. L., Davis M. M. Polymerase chain reaction with single-sided specificity: analysis of T cell receptor delta chain. Science. 1989 Jan 13;243(4888):217–220. doi: 10.1126/science.2463672. [DOI] [PubMed] [Google Scholar]
- Majumder A. L., Akins R. A., Wilkinson J. G., Kelley R. L., Snook A. J., Lambowitz A. M. Involvement of tyrosyl-tRNA synthetase in splicing of group I introns in Neurospora crassa mitochondria: biochemical and immunochemical analyses of splicing activity. Mol Cell Biol. 1989 May;9(5):2089–2104. doi: 10.1128/mcb.9.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R. M., Pleij C. W., Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur J Biochem. 1991 Oct 15;201(2):303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- McClure M. A. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991 Nov;8(6):835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Bujarski J. J., Dreher T. W., Hall T. C. Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986 Feb 20;187(4):537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Nargang F. E., Bell J. B., Stohl L. L., Lambowitz A. M. The DNA sequence and genetic organization of a Neurospora mitochondrial plasmid suggest a relationship to introns and mobile elements. Cell. 1984 Sep;38(2):441–453. doi: 10.1016/0092-8674(84)90499-9. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville B. J., Collins R. A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990 May 18;61(4):685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- Schulte U., Lambowitz A. M. The LaBelle mitochondrial plasmid of Neurospora intermedia encodes a novel DNA polymerase that may be derived from a reverse transcriptase. Mol Cell Biol. 1991 Mar;11(3):1696–1706. doi: 10.1128/mcb.11.3.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Maizels N. tRNA-like structures tag the 3' ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Gallo R. C. Reverse transcriptase. CRC Crit Rev Biochem. 1975 Dec;3(3):289–347. doi: 10.3109/10409237509105454. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]