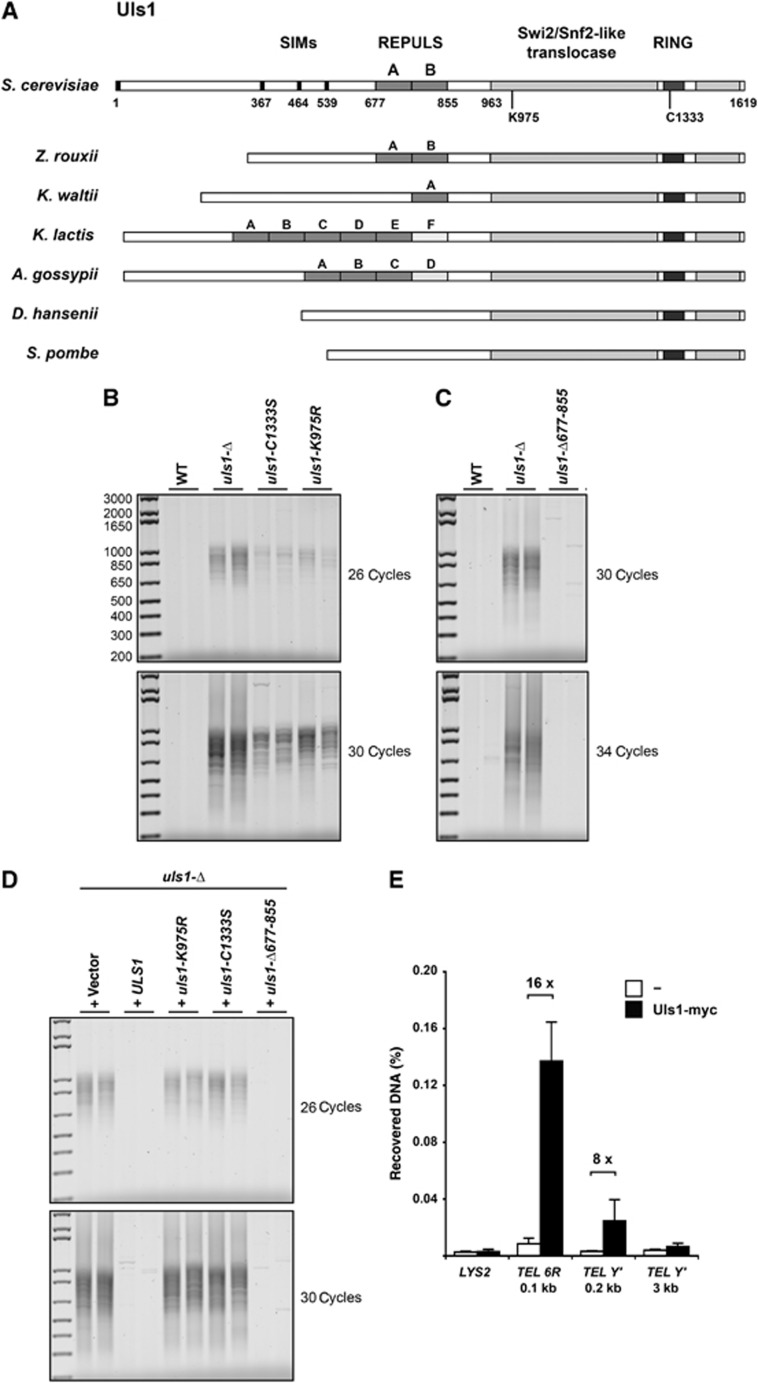
Figure 2.
Mutations in Uls1 translocase and ubiquitin ligase catalytic domains cause telomere fusions. (A) Schematic representation of S. cerevisiae Uls1 and Uls1 from other representative yeast species. REPULS domains are only present in Saccharomycetaceae, in tandem, isolated or in multiple copies, some of which being degenerated (light grey, see the multiple alignment in Supplementary Figure S1). REPULS domains are not present in hypothetical proteins likely corresponding to the Uls1 orthologues in D. hansenii (UniProt Identifier Q6BMG3) and in S. pombe (two hypothetical orthologues: UniProt Identifier O60177 and O13762). (B) Telomere fusions occur in uls1-C1333S and uls1-K975R cells. Strains 206-2b and 205-9a (WT), 206-1d and 205-14c (uls1-Δ), 199-3a (uls1-C1333S) and 200-2c and 200-5d (uls1-K975R) were grown to stationary phase. Telomere fusions were amplified with 26 and 30 cycles. (C) Telomere fusions are undetectable in usl1-Δ677-855 cells. Strains 196-11c and 196-13b (WT), 196-5a and 196-6a (uls1-Δ) and Lev791 and Lev792 (uls1-Δ677-855) were grown to stationary phase. Telomere fusions were amplified with 30 and 34 cycles. (D) Complementation of uls1-Δ by ULS1 wild-type and mutant alleles. Strain 210-5b (uls1-Δ) was transformed with StuI-digested plasmids pRS406, pRS406-uls1-K975R, pRS406-uls1-C1333S and pRS406-uls1-Δ677-855 and with MfeI-digested plasmid pRS404-ULS1. Cultures of two independent transformants were grown to stationary phase in rich medium. Telomere fusions were amplified with 26 and 30 cycles. (E) Uls1 is present at telomeres. Uls1 was tagged with 13 myc epitopes in the C-terminal position (Uls1-myc). A wild-type strain with untagged Uls1 was used as a control (−). Distances of the primer pairs from the terminal TG1–3 repeats are indicated. LYS2 is an internal non-telomeric locus. ChIP assay was performed as described by Guglielmi et al (2007). Immunoprecipitated and input DNA were quantified by qPCR. ChIP signal was normalized to input DNA. Data shown are the mean and standard deviation of three independent experiments.