Figure 7.
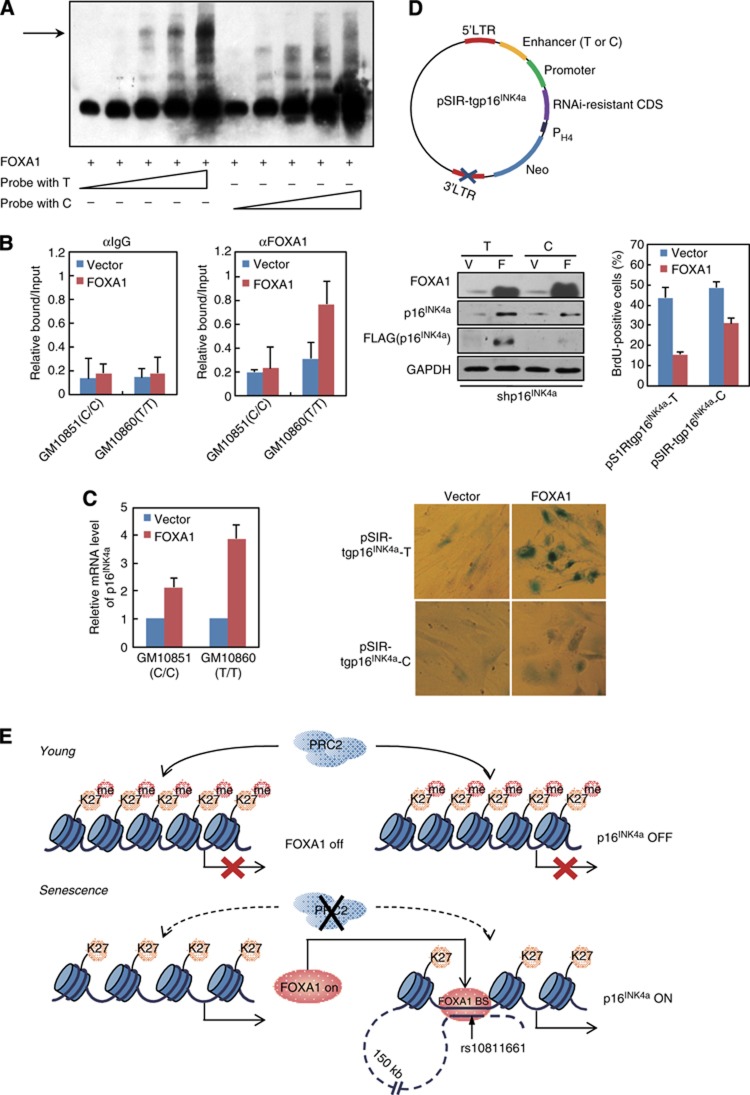
DNA variant at the −150 kb enhancer influences FOXA1-dependent p16INK4a activation. (A) FOXA1 possessed higher affinity towards T-allele binding site located in the −150 kb enhancer. EMSA was performed with recombinant FOXA1 protein and increasing amount of biotin labelled probe. The probe harbours a DNA variant rs10811661 for T or C, which is enclosed in the predicted FOXA1 binding motif at the −150 kb enhancer. (B) ChIP analysis of FOXA1 enrichment at the −150 kb enhancer was performed in FOXA1- or control vector-infected GM10851 (carrying C allele) and GM10860 (carrying T allele) lymphocytes. Data represent the mean+s.d. for triplicate experiments. (C) GM10851 and GM10860 lymphocytes were transduced with lentiviral FOXA1 or control vector. RT-qPCR analysis for p16INK4a was conducted in these cells. Data represent the mean+s.d. for triplicate experiments. (D) A transgenic construct designated as pSIR-tgp16IN4a hosted on a self-inactivating retrovirus-derived vectors was demonstrated in the top panel. RNAi-resistant CDS, FLAG-tagged full-length p16INK4a coding sequence with synonymous mutations to abolish its pairing with shp16INK4a; promoter, p16INK4a promoter with 370 bp DNA sequence upstream from its TSS; enhancer (T or C), −150 kb enhancer with DNA variant rs10811661 for T or C. PH4, promoter for histone H4. Early-passage 2BS cells with endogenous p16INK4a silenced by its targeting shRNA (shp16INK4a) were co-transduced with pSIR-tgp16INK4a (T or C) and retroviral FOXA1 or control vector. The cells were selected and cultured for 2 weeks. Western blotting analysis was performed with indicated antibodies (middle panel, left). Phenotypic differences in these cells were monitored through BrdU incorporation assay (middle panel, right) and SA-β-gal activity analysis (bottom panel). Data represent the mean+s.d. for triplicate experiments. (E) A model depicts the molecular mechanism underlying FOXA1-mediated p16INK4a activation during senescence.
Source data for this figure is available on the online supplementary information page.