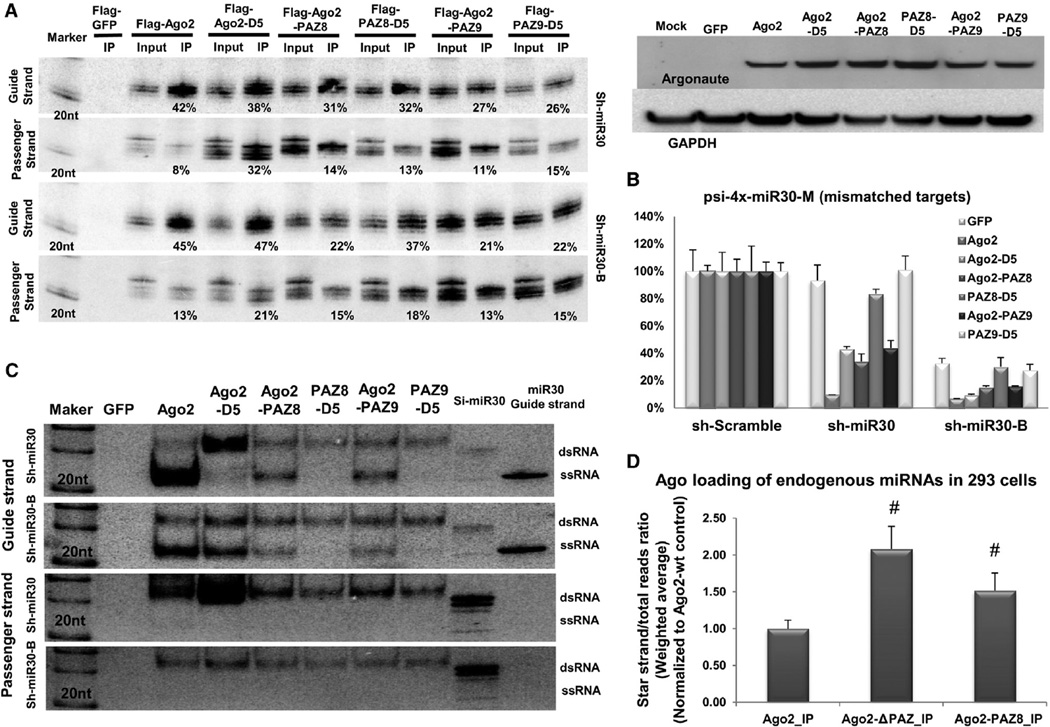
Figure 4. Ago2 with Point Mutations in PAZ Is Defective in Slicer-Independent RISC Unwinding.
(A) Sh-miR30 or shRNA-miR30-B coexpressed with Flag-GFP or various Flag-Ago2 mutants in Ago2 KO MEF cells. IP and northern blotting experiments were performed as described in Figure 1. The expression levels of the proteins used in the assays were assessed by western blotting with Ago2 antibody. The signal from GAPDH served as the transfection and loading controls.
(B) A psiCHECK vector with four tandem target sites in the 3′ UTR, which were mismatched to the guide strand of sh-miR30 and sh-miR30-B, was cotransfected with various shRNAs and Argonaute-expressing plasmids in Ago2 KO MEF cells. Dual-luciferase assays were performed and analyzed as described in Figure 1. Error bars represent the SD from two independent experiments, each performed in triplicate transfections.
(C) IP samples in (A) were separated on 15% polyacrylamide native gels. 20 fmol synthetic RNAs (si-miR30 duplex and single-stranded guide-strand RNA of miR-30) were also loaded on the gel as controls. Guide- and passenger-strand RNAs were identified by sequential northern blot. Notably, the si-miR30 control ran as a mixture of dsRNA and a trace amount of ssRNA.
(D) The amount of miRNAs and their corresponding star strands were measured in various Ago-IP samples by deep sequencing. For each miRNA, the percentage of its star strand to total reads was determined. The weighted average and SD were calculated by using the total reads of each miRNA as the weight. Number sign (#) indicates statistical significance (p < 0.05, unpaired t test, using Ago2-IP as a control group). See also Figure S4 and Table S1.