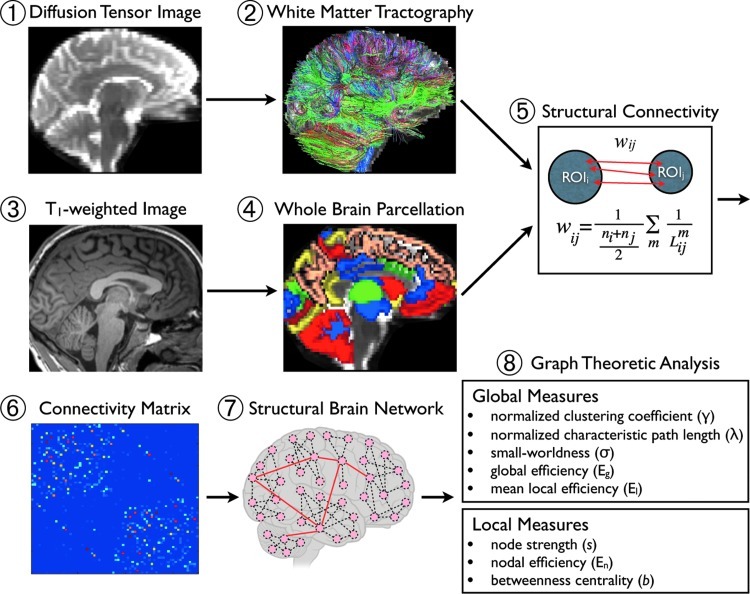
FIG. 1.
Overview of the network analysis for structural brain connectivity. For each subject, diffusion tensor images (1) were used to generate the white matter fiber tracts (2) within the whole brain using streamline tractography with 30 random seeds in a voxel. The individual T1-weighted image (3) was divided into 88 ROIs, (4) including cerebral cortex, subcortex, and cerebellum, using FreeSurfer. The T1-weighted image was co-registered to the corresponding non-diffusion-weighted image using 12-df parameter affine transformation, and the transformation matrix was applied to each ROI to generate corresponding volumes in diffusion native space. Structural connectivity (5) was defined by the number and length of fibers connecting regions i and j. Then, a weighted connectivity matrix (6) generated the structural brain network (7). Finally, the global and local network measures (8) were computed and compared between CB subjects and healthy controls. ROI, regions of interest; CB, cannabis.