Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data set | Apo form | Hg derivative (peak) | Meropenem complex |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | P212121 |
| Unit-cell parameters | |||
| a (Å) | 135.6 | 135.7 | 68.9 |
| b (Å) | 58.6 | 58.4 | 73.4 |
| c (Å) | 40.9 | 41.0 | 104.1 |
| α = γ (°) | 90.0 | 90.0 | 90.0 |
| β (°) | 94.4 | 94.3 | 90.0 |
| X-ray wavelength (Å) | 1.00000 | 1.00600 | 1.00000 |
| Resolution range (Å) | 20.0–1.80 (1.83–1.80) | 50.0–1.79 (1.82–1.79) | 50.0–2.00 (2.03–2.00) |
| Total No. of reflections | 114133 (3344) | 218163 (7927)† | 254969 (11833) |
| No. of unique reflections | 29426 (1045) | 58915 (2557)† | 34693 (1621) |
| Completeness (%) | 97.9 (70.4) | 99.3 (86.8)† | 95.5 (91.2) |
| 〈I/σ(I)〉 | 46.5 (9.1) | 43.0 (7.4)† | 31.0 (4.3) |
| R merge ‡ (%) | 4.6 (14.4) | 4.6 (14.9)† | 12.8 (89.1) |
| SAD phasing | |||
| Figure of merit (before/after density modification) | 0.41/0.60 | ||
| Model refinement | |||
| PDB code | 4gsq | 4gsr | 4gsu |
| Resolution range (Å) | 20.0–1.80 | 20.0–1.79 | 20.0–2.00 |
| R work/R free § (%) | 19.7/23.3 | 19.6/22.6 | 18.9/23.2 |
| No. of non-H atoms/average B factor (Å2) | |||
| Protein | 1889/23.5 | 2014/23.1 | 4112/26.4 |
| Water oxygen | 206/32.9 | 230/36.3 | 228/33.0 |
| Meropenem | — | — | 52/65.5 |
| EMTS | — | 11/38.0 | — |
| Glycerol | 6/53.3 | 12/44.1 | — |
| Calcium ion | 1/65.2 | — | — |
| Wilson B factor (Å2) | 21.9 | 20.4 | 23.5 |
| R.m.s. deviations from ideal geometry | |||
| Bond lengths (Å) | 0.009 | 0.007 | 0.010 |
| Bond angles (°) | 1.29 | 1.15 | 1.40 |
| R.m.s. Z-scores¶ | |||
| Bond lengths | 0.450 | 0.335 | 0.491 |
| Bond angles | 0.590 | 0.512 | 0.622 |
| Ramachandran plot†† (%) | |||
| Favoured/outliers | 98.8/0.0 | 98.5/0.0 | 97.9/0.2 |
| Poor rotamers†† (%) | 1.00 | 0.47 | 0.91 |
Friedel pairs were treated as separate observations.
R
merge = 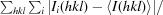
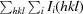 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl, 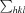 is the sum over all reflections and
is the sum over all reflections and 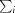 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
R
work = 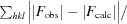
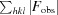 , where R
free is calculated for a randomly chosen 5% of reflections which were not used for structure refinement and R
work is calculated for the remaining reflections.
, where R
free is calculated for a randomly chosen 5% of reflections which were not used for structure refinement and R
work is calculated for the remaining reflections.
Values obtained using REFMAC.
Values obtained using MolProbity.
