Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| LpPgb1 | Apo LpPgb1 | SmBgl | SmBglBG6 | E375Q SmBglPSC | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P622 | P31 | P21 | P21 | P41212 |
| Unit-cell parameters (, ) | a = 150.6, c = 95.9 | a = 96.1, c = 289.1 | a = 58.9, b = 91.0, c = 98.6, = 98.7 | a = 58.7, b = 92.4, c = 94.4, = 101.3 | a = 82.3, c = 221.2 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 |
| Radiation source | APS ID-19 | APS BM-19 | APS ID-19 | APS ID-19 | APS ID-19 |
| Wavelength () | 0.9792 | 0.9790 | 0.9794 | 0.9792 | 0.9793 |
| Resolution () | 50.001.50 (1.531.50) | 50.002.31 (2.352.31) | 36.001.70 (1.731.70) | 25.101.48 (1.511.48) | 39.002.54 (2.592.54) |
| Unique reflections | 102382 | 130353 | 112307 | 163713 | 25874 |
| R merge † | 0.112 (0.650) | 0.079 (0.285) | 0.115 (0.710) | 0.067 (0.589) | 0.087 (0.636) |
| I/(I) | 33.2 (3.7) | 21.8 (2.75) | 26.8 (1.9) | 27.1 (1.9) | 25.8 (2.3) |
| Completeness (%) | 99.9 (100) | 99.6 (96.7) | 98.6 (97.2) | 99.7 (96.5) | 99.4 (100) |
| Multiplicity | 11.7 (11.4) | 4.4 (2.8) | 4.1 (4.0) | 4.6 (3.1) | 6.7 (6.8) |
| Refinement | |||||
| Resolution () | 31.001.50 | 30.02.31 | 35.901.69 | 25.101.48 | 39.002.54 |
| Reflections (work/test set) | 101314/1031 | 129000/1248 | 100643/5294 | 155447/8214 | 24479/1312 |
| R work/R free ‡ | 0.118/0.134 | 0.174/0.213 | 0.163/0.193 | 0.162/0.185 | 0.180/0.229 |
| No. of atoms | |||||
| Protein | 3993 | 23032 | 7846 | 7925 | 3902 |
| Ligands | 36 | 12 | 164 | 58 | 36 |
| Water | 589 | 547 | 565 | 1145 | 34 |
| Average B factor (2) | |||||
| Protein | 12.7 | 42.1 | 37.3 | 18.5 | 69.4 |
| Ligands | 28.2 | 52.7 | 60.4 | 23.8 | 79.0 |
| Water | 25.9 | 38.9 | 39.5 | 30.8 | 57.6 |
| R.m.s. deviations from ideal§ | |||||
| Bond lengths () | 0.015 | 0.013 | 0.006 | 0.006 | 0.007 |
| Bond angles () | 1.455 | 1.302 | 0.939 | 1.060 | 1.058 |
| Clashscore [percentile]¶ | 1.84 [99th] | 8.13 [97th] | 9.25 [73rd] | 5.36 [91st] | 13.08 [91st] |
| Poor rotamers¶ (%) | 0.75 | 1.85 | 1.99 | 1.74 | 4.98 |
| Ramachandran statistics of / angles¶ (%) | |||||
| Most favored | 99.2 | 97.5 | 97.7 | 98.3 | 97.5 |
| Outliers | 0 | 0 | 0 | 0 | 0 |
| PDB entry | 3qom | 4gze | 3pn8 | 4f66 | 4f79 |
R
merge = 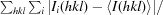
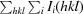 , where I
i(hkl) is the intensity of observation i of reflection hkl.
, where I
i(hkl) is the intensity of observation i of reflection hkl.
R
work = 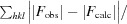
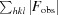 for all reflections, where F
obs and F
calc are observed and calculated structure factors, respectively. R
free is calculated analogously for the test reflections, which were randomly selected and excluded from the refinement.
for all reflections, where F
obs and F
calc are observed and calculated structure factors, respectively. R
free is calculated analogously for the test reflections, which were randomly selected and excluded from the refinement.
According to Engh and Huber parameters (Engh Huber, 1991 ▶).
According to MolProbity (Chen et al., 2010 ▶).
