Table 3. Posterior nodes and standard deviations of mixture model parameters describing resistance in treatment-experienced patients.
| Region | Tree weights comprising measure of similarity | Mixture model parameters from trees unique to treatment-experienced patients | |||||||||
| AFR |
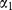
|
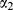
|
WT H H |
WT R R |
WT E E |
WT S S |
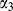
|
WT H H |
H R R |
R E E |
H S S |
| 0.52(0.18) | 0.20(0.015) | 0.47(0.053) | 0.13(0.029) | 0.062(0.012) | 0.35(0.036) | 0.27(0.18) | 0.34(0.240) | 0.84(0.097) | 0.57(0.079) | 0.68(0.065) | |
| AMR |
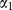
|
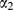
|
WT H H |
WT R R |
WT E E |
WT S S |
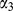
|
WT H H |
H R R |
S E E |
H S S |
| 0.71(0.032) | 0.20(0.011) | 0.62(0.032) | 0.47(0.031) | 0.11(0.014) | 0.21(0.029) | 0.088(0.030) | 0.69(0.190) | 0.94(0.033) | 0.59(0.058) | 0.95(0.045) | |
| EMR |
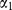
|
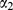
|
WT H H |
WT R R |
WT E E |
WT S S |
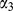
|
S H H |
H R R |
R E E |
WT S S |
| 0.36(0.120) | 0.24(0.028) | 0.58(0.069) | 0.58(0.069) | 0.22(0.045) | 0.55(0.066) | 0.40(0.120) | 0.87(0.055) | 0.95(0.034) | 0.78(0.073) | 0.58(0.160) | |
| FSU |
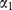
|
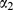
|
WT H H |
WT R R |
WT E E |
WT S S |
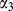
|
WT H H |
H R R |
R E E |
H S S |
| 0.33(0.034) | 0.24(0.014) | 0.67(0.028) | 0.31(0.031) | 0.25(0.020) | 0.57(0.020) | 0.43(0.032) | 0.93(0.048) | 0.97(0.018) | 0.74(0.015) | 0.71(0.011) | |
| NFSU-EUR |
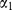
|
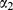
|
WT H H |
WT R R |
WT E E |
WT S S |
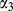
|
WT H H |
H R R |
R E E |
H S S |
| 0.48(0.180) | 0.18(0.010) | 0.55(0.039) | 0.19(0.030) | 0.10(0.016) | 0.34(0.031) | 0.34(0.180) | 0.25(0.190) | 0.97(0.030) | 0.64(0.055) | 0.85(0.045) | |
| SEAR |
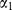
|
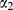
|
WT H H |
WT R R |
WT E E |
WT S S |
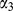
|
S H H |
H R R |
R E E |
WT S S |
| 0.29(0.12) | 0.22(0.033) | 0.92(0.027) | 0.38(0.091) | 0.11(0.045) | 0.37(0.057) | 0.25(0.100) | 0.50(0.200) | 0.80(0.200) | 0.76(0.220) | 0.50(0.210) | |
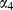
|
WT H H |
H R R |
R E E |
H S S |
|||||||
| 0.24(0.099) | 0.40(0.210) | 0.66(0.260) | 0.74(0.180) | 0.47(0.270) | |||||||
| WPR |
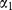
|
|
WT H H |
WT R R |
WT E E |
WT S S |
|
WT H H |
H R R |
R E E |
H S S |
| 0.51(0.056) | 0.30(0.016) | 0.67(0.029) | 0.46(0.033) | 0.12(0.014) | 0.41(0.020) | 0.19(0.053) | 0.68(0.160) | 0.93(0.049) | 0.89(0.069) | 0.60(0.028) | |
 corresponds to the tree weights of the prespecified trees. Nodes = {WT = wild type, H = isoniazid, R = rifampin, E = ethambutol and S = streptomycin }.
corresponds to the tree weights of the prespecified trees. Nodes = {WT = wild type, H = isoniazid, R = rifampin, E = ethambutol and S = streptomycin }.
